INTRODUCTION
Anemia is caused by abnormal hematopoietic cells, lack of vitamins and minerals in red blood cells, high vitamin demand due to pregnancy and lactation, and chronic bleeding due to digestive disease [1]. Clinically, anemia can cause general fatigue, hair loss, and mental illness. Moreover, the burden on the heart due to the lack of hemoglobin is related to cardiomyopathy [2]. While the prevalence of anemia is increasing annually in developed countries, the increase in anemia-related deaths is a problem that cannot been neglected in recent years [3]. Chronic iron deficiency can lead to anemia as iron is a cofactor for physiological proteins and is indispensable for life cycle activities, such as carrying oxygen to the mitochondria [4]. Some iron-regulating factors in the small intestinal epithelium, such as hepatocytes, macrophages, and the bone marrow, regulate iron homeostasis [5]. Dietary ferric (Fe3+) is reduced to ferrous (Fe2+) by duodenal cytochrome B, located at the duodenal brush border membrane. Subsequently, Fe2+ passes through divalent metal transporter 1, which is expressed intracellularly at the surface cells [6, 7]. Alternatively, when there is a high hepcidin or low ferritin level in the epithelium, Fe2+ is oxidized again and stored in the epithelium as ferritin. Ferrous ions pass through ferroportin, which is the only channel to release iron into the blood flow. These ferrous ions get oxidized by hephaestin located at the basement membrane and then iron is transported to various organs [8].
Hepcidin, encoded by Hamp, is a peptide hormone produced and secreted by hepatocytes that influences duodenal epithelial cells and regulates dietary iron uptake. More specifically, hepcidin is a downregulatory factor that inhibits iron intake by binding with ferroportin and promoting the disassembly of ferroportin in the lysosome [9]. The function of hepcidin and the relationship between its controlling factors have been well assessed in previous studies [10]. One of the heme-binding proteins, known as progesterone receptor membrane component 1 (Pgrmc1), is a non-canonical progesterone receptor associated with diverse molecular gene regulation. Pgrmc1 functions in a variety of cellular processes, including heme binding, drug metabolism, progesterone responsiveness, steroidogenesis, female fertility, lipid transport, insulin secretion, cancer, and anti-apoptosis [11–13]. When its expression is substantially higher in the liver, it might be contained that hepatic Pgrmc1 controls iron homeostasis. According to a previous study, progesterone increases Hamp mRNA expression by increasing Pgrmc1 in zebrafish and HepG2 cells [14]. While Pgrmc1 can regulate Hamp mRNA expression, the role of Pgrmc1 has not been studied in a mammalian model. Thus, to investigate whether Pgrmc1 regulates hepcidin in mice, we generated Pgrmc1 knockout (KO) mice and conduct molecular biological analyses using livers.
MATERIALS AND METHODS
C57BL/6J wild-type (WT) and Pgrmc1 KO mice (both 10-week-old; male) were housed in a temperature- and light-controlled facility at Chungnam National University and fed standard chow with water provided ad libitum. All mouse experiments were approved and performed in accordance with the guidelines of the Chungnam Facility Animal Care Committee (202209A-CNU-192). Before necropsy, WT or Pgrmc1 KO mice were fasted for 18 hr and then fed for 5 hr under a general feeding regimen.
RNA was isolated using the TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), chloroform (Sigma-Aldrich, St. Louis, MO, USA), and isopropanol (Kanto Chemical, Tokyo, Japan) dissolved in water treated with diethyl pyrocarbonate (DEPC; Amresco, Solon, OH, USA). cDNA was acquired using a Reverse Transcriptase Kit (SmartGene, Lausanne, Switzerland). Specific primers, SYBR Green Q-PCR Master Mix with Low Rox (SmartGene), and Stratagene Mx3000P (Agilent Technologies, Santa Clara, CA, USA) were used to perform real-time PCR.
The human primers used were as follows: PGRMC1 forward (AAA GGC CGC AAA TTC TAC GG), PGRMC1 reverse (CCC AGT CAC TCA GAG TCT CCT), Hamp forward (TGA CCA GTG GCT CTG TTT TC), Hamp reverse (GAA AAT GCA GAT GGG GAA GT), RPLP0 forward (TCG ACA ATG GCA GCA TCT AC), and RPLP0 reverse (GCC TTG ACC TTT TCA GCA AG).
The mouse primers used were as follows: Pgrmc1 forward (GGC AAG GTG TTC GAC GTG A), Pgrmc1 reverse (GTC CAG GCA AAA TGT GGC AA), Hamp forward (CTG CCT GTC TCC TGC TTC TC), Hamp reverse (AGA TGC AGA TGG GGA AGT TG), Fth1 forward (CGA GAT GAT GTG GCT CTG AA), Fth1 reverse (GTG CAC ACT CCA TTG CAT TC), Ftl1 forward (GGG CCT CCT ACA CCT ACC TC), Ftl1 reverse (CTC CTG GGT TTT ACC CCA TT), Bmp6 forward (TTC TTC AAG GTG AGC GAG GT), Bmp6 reverse (TAG TTG GCA GCG TAG CCT TT), IL-6 forward (CTG CAA GAG ACT TCC ATC CAG), IL-6 reverse (AGT GGT ATA GAC AGG TCT GTT GG), Rplp0 forward (GCA GCA GAT CCG CAT GTC GCT CCG), and Rplp0 reverse (GAG CTG GCA CAG TGA CCT CAC ACG G).
Liver, primary hepatocytes, and HepG2 cells were lysed in T-PER buffer (Thermo Fisher Scientific) and sonicated for protein extraction. Protein samples were quantified using the protein measurement solution (iNtRON, Seongnam, Korea) and ran in a 10%–12% SDS-PAGE after 5 min of boiling at 100°C. The gels were blotted at 300 mA for 80 min. PVDF membranes were blocked in 3% bovine serum albumin (BSA) for 15 min and incubated with primary antibodies diluted 1:2,500 in 3% BSA at 4°C for overnight. The following day, membranes were incubated with secondary antibodies diluted 1:2,500 in 5% skim milk at 4°C for overnight. The results were detected using ECL solution (Cyanagen, Bologna, Italy). The primary antibodies used were rabbit anti-Pgrmc1, rabbit anti-p38, rabbit anti-phospho-p38, rabbit anti-Akt, rabbit anti-phospho-Akt, rabbit anti-Hsp90 (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-hepcidin, rabbit anti-Stat3, rabbit anti-phospho-Stat3, rabbit anti-Creb, and rabbit anti-phospho-Creb (ABclonal, Shanghai, China).
HepG2 cells obtained from the Korean Cell Line Bank. All the cell culture reagents were purchased from Welgene (Gyeongsan, Korea). HepG2 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 5% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/mol), streptomycin (100 µg/mL), ciprofloxacin (10 µg/mL), and gentamycin (50 µg/mL) at 37°C in 5% CO2. HepG2 cells were washed with Dulbecco’s Phosphate-Buffered Saline (DPBS) and incubated in a high-glucose medium (4,500 mg/dL, without FBS) for 3 hr. Forskolin was added for 30 min or 1 hr after determining the treatment concentration.
For PGRMC1 knockdown, HepG2 cells were transfected with negative control siRNA or PGRMC1 siRNA and Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) in Opti-MEM (Thermo Fisher Scientific) medium for 72 hr. PGRMC1 siRNA #1 (5'-CAGUACAGUCGCUAGUCAA-3') and #2 (5'-CAGUUCACUUUCAAGUAUCA-U-3') were purchased from Bioneer (Daejeon, Korea).
Primary hepatocytes were maintained in a DMEM with 10% FBS, penicillin (100 U/mol) and streptomycin (100 µg/mL) at 37°C in 5% CO2. Hepatocytes were washed with DPBS and incubated in high-glucose medium or high glucose and 10 nM insulin for 3 hr before harvest.
Primary hepatocytes were isolated from WT and Pgrmc1 KO mice. The animals were anesthetized with 2% isoflurane and 1% O2, perfused with Ca2+ and Mg2+ free-Hanks Balanced Salt Solution (HBSS) containing 0.5 mM EDTA and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and then digested with HBSS containing Ca2+ and Liberase (Sigma-Aldrich). The livers were removed, rinsed with phosphate-buffered saline (PBS), and loosened in DMEM. After washing a few times, primary hepatocytes were seeded in cell culture plates and cultured in DMEM supplemented with 10% heat-inactivated FBS, penicillin (100 U/mol), and streptomycin (100 µg/mL) at 37°C in 5% CO2. Hepatocytes were treated with 10 nM insulin for 3 hr after incubation in a high-glucose DMEM (containing 4,500 mg/dL glucose) without FBS for 3 hr.
RESULTS
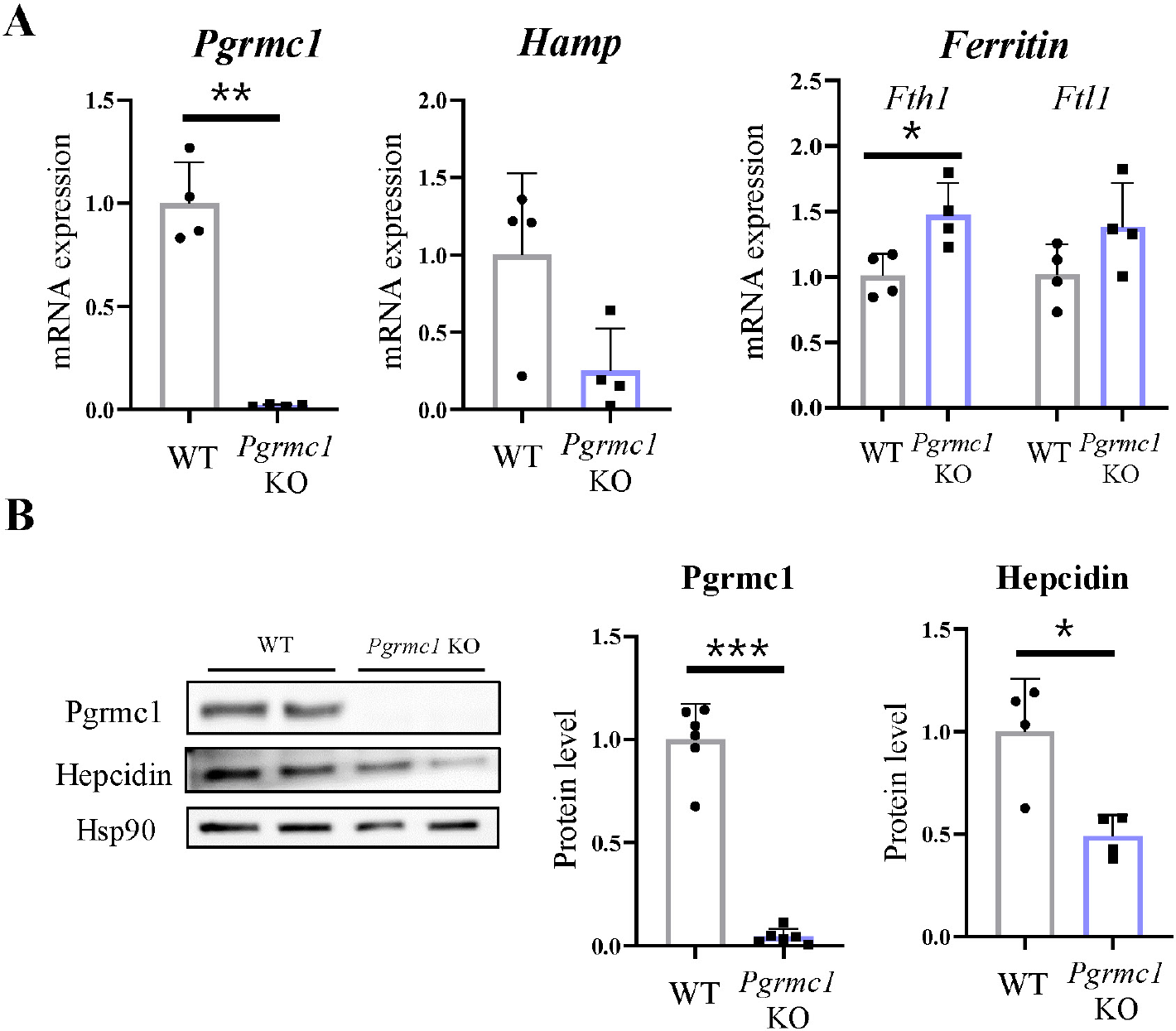
To investigate the relationship between Pgrmc1 and hepcidin, male WT or Pgrmc1 KO mice (n = 4) were observed. The mice were fasted for 18 hr and then fed a regular diet for 5 hr before necropsy. In terms of food intake status, Pgrmc1 KO mice showed lower Hamp mRNA expression in the liver than WT mice. Ferritin heavy chain 1 (Fth1) and ferritin light chain 1 (Ftl1) are the major intracellular iron storage proteins. Their gene expression levels were lower than those in the WT mice (Fig. 1A). Moreover, hepcidin protein levels in Pgrmc1 KO mice were lower than those in WT mice, suggesting that Pgrmc1 regulates hepcidin levels in vivo (Fig. 1B).
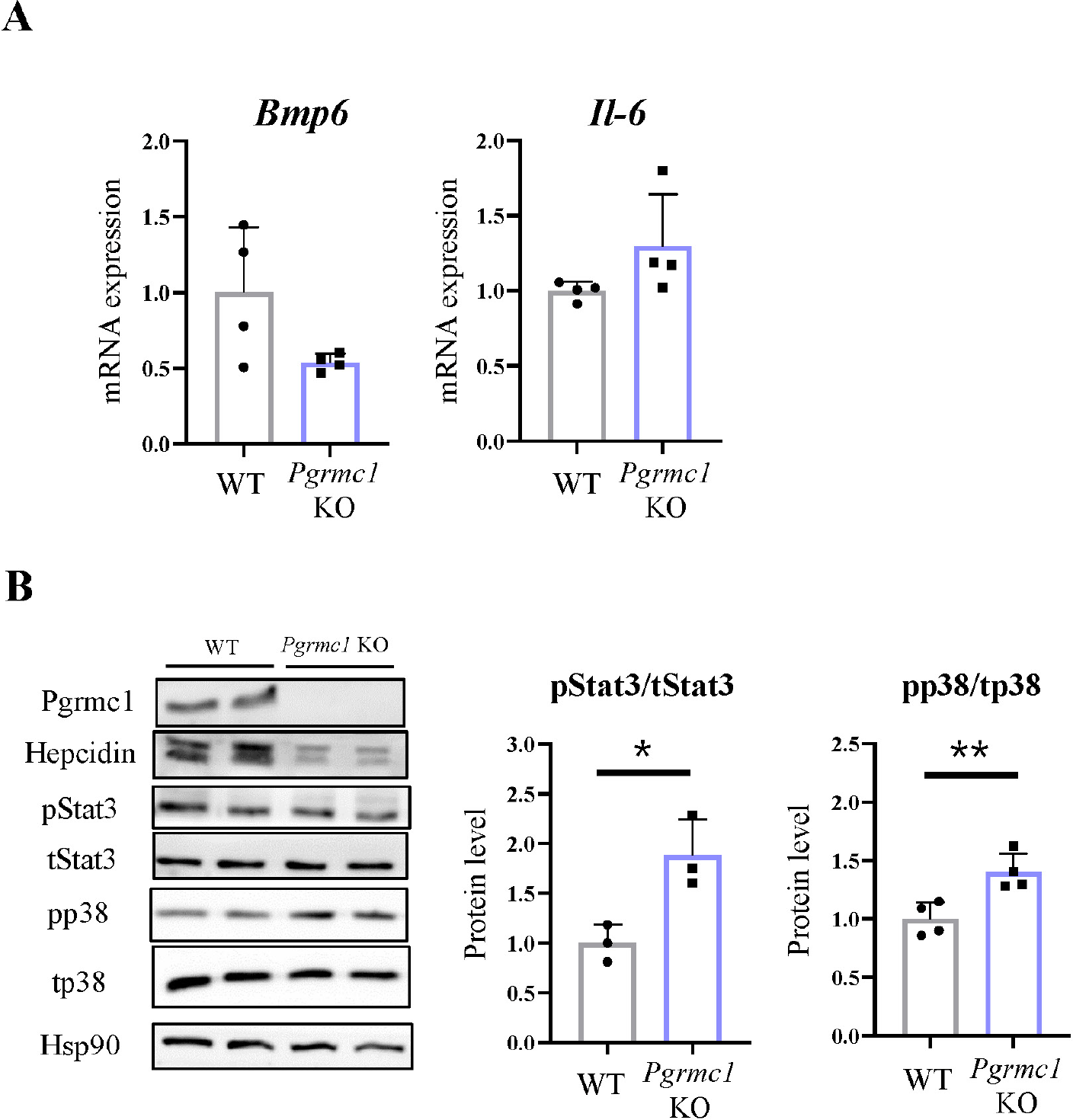
Hepcidin is regulated by the BMP-SMAD pathway or by an increase in inflammatory mediators, such as IL-6, JAK, and STAT3 signaling [15]. To determine how Pgrmc1 regulates hepcidin in the liver, we measured Bmp6 and IL-6 mRNA expression using qPCR. However, no significant differences were found between WT and Pgrmc1 KO mice (Fig. 2A). Furthermore, the ratio of phospho-Stat3, an inflammatory marker expressed by IL-6 in hepcidin transcription signaling, was lower in WT mice than in Pgrmc1 KO mice (Fig. 2B).
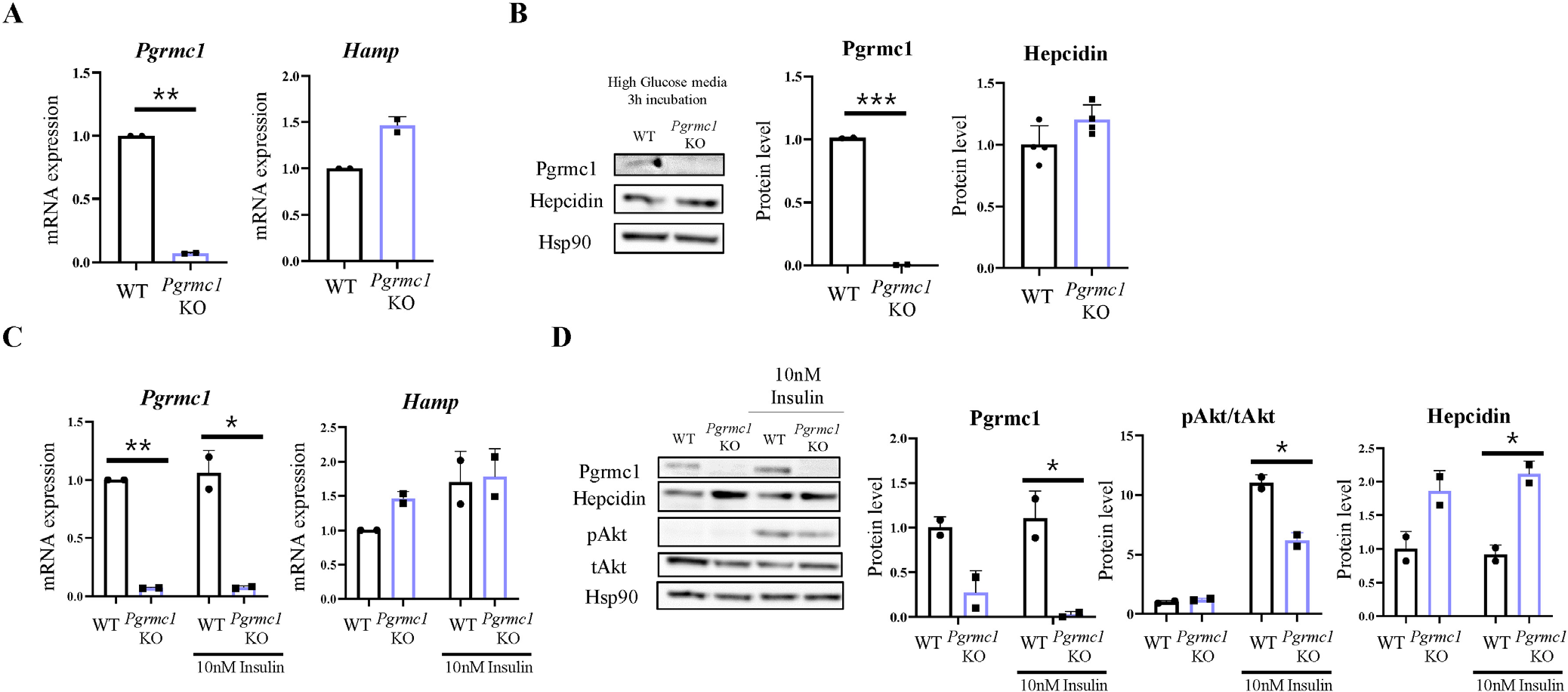
To determine the factors that regulate hepcidin via Pgrmc1, we isolated primary hepatocytes from mice. The primary hepatocytes were incubated in high-glucose DMEM containing penicillin (100 U/mol), streptomycin (100 µg/mL), and 10% FBS for 24 hr. To assess the difference between primary hepatocytes of WT and Pgrmc1 KO mice, we introduced primary hepatocytes into a high-glucose DMEM without FBS for 3 hr. However, no difference was found in hepcidin protein levels and Hamp mRNA expression between WT and Pgrmc1 KO mice (Fig. 3A and B). Next, primary hepatocytes from WT and Pgrmc1 KO mice were treated with 10 nM insulin for 3 hr after incubation in a high-glucose DMEM without FBS for 3 hr. While the ratio of phospho-Akt levels increased in the insulin-treated group, the hepcidin protein level failed to increase in the insulin-treated group. When Hamp mRNA expression was higher in the insulin-treated group, no difference was found between the WT and Pgrmc1 KO mice (Fig. 3C). Additionally, Pgrmc1 KO mice showed higher hepcidin protein levels than WT mice in each group (Fig. 3D). These results suggest that Pgrmc1 is not related to the hepcidin-glucose or hepcidin-insulin responses.
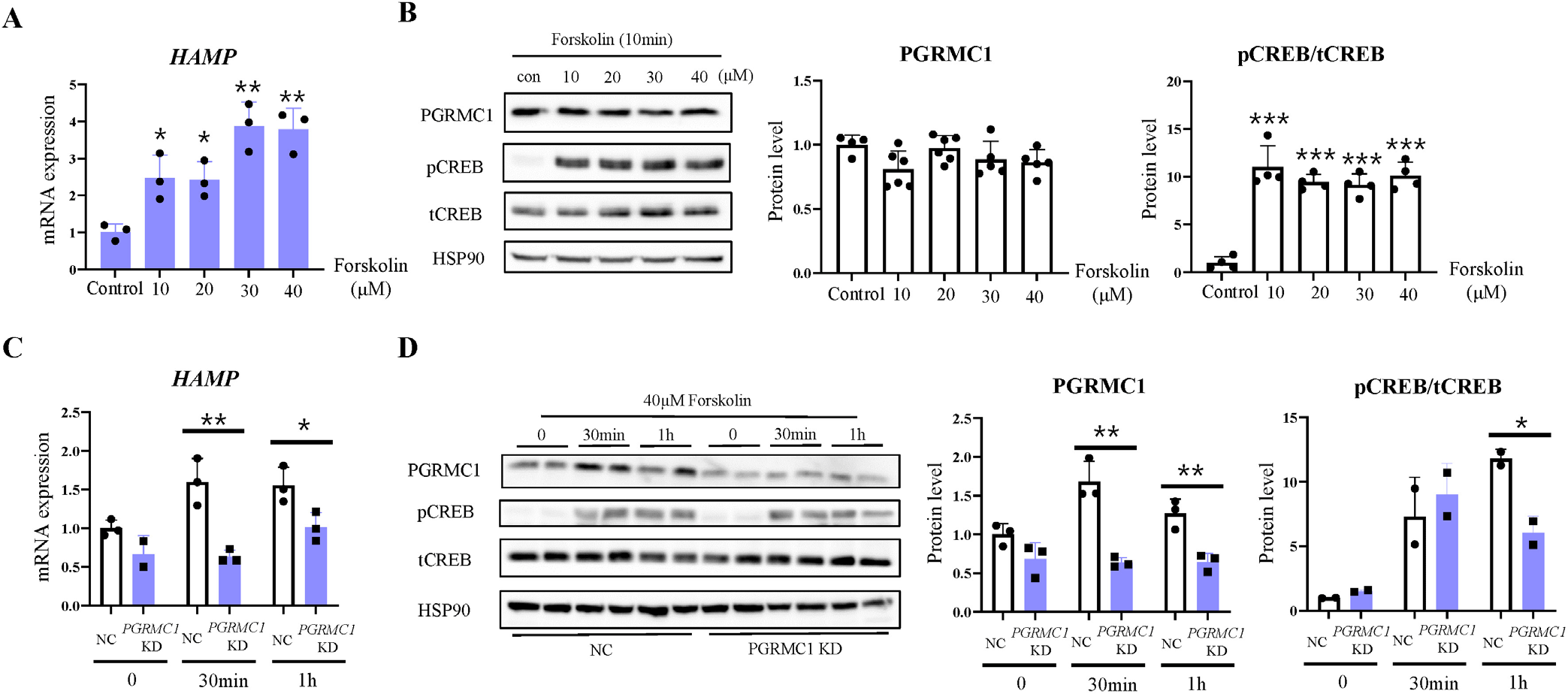
As phosphorylation of cAMP-response element-binding protein (Creb) increases Hamp transcription [16] and Pgrmc1 increases cAMP activation during fasting [17], it is possible that Pgrmc1 controls Hamp mRNA transcription via cAMP activation. To determine whether the cAMP activator increased Hamp mRNA levels, we treated HepG2 cells with forskolin, which is a substitute for glucagon. CREB was phosphorylated by forskolin treatment. As the forskolin concentration increased in a dose-dependent manner, HAMP mRNA levels also significantly increased (Fig. 4A and B). When PGRMC1 was knocked down by siPGRMC1 in HepG2 cells, HAMP mRNA showed low reactivity against forskolin compared to the negative control (NC), suggesting that PGRMC1 promotes HAMP mRNA transcription via cAMP activation (Fig. 4C and D).
DISCUSSION
Hepcidin is a critical limiting factor for iron absorption. Iron deficiency anemia is the most prevalent nutritional disorder worldwide, and its risk is higher among women who experience periods that are especially long or that include very heavy bleeding [3]. Women have lower iron levels than men, and serum pro-hepcidin levels are significantly higher in men than in healthy women [18]. However, girls show higher serum hepcidin levels than boys [19]. These reports suggest that serum hepcidin levels may be affected by iron depletion, including ongoing blood loss, recent pregnancy, and inadequate dietary iron intake. Notably, female laboratory rats that did not experience menstrual bleeding showed higher hepatic hepcidin levels than male rats [20]. Chronic low iron concentrations may affect hepcidin levels more than other factors. Therefore, clarification of the mechanism of hepcidin regulation using physiological approaches is necessary to resolve the instability of iron homeostasis.
Some studies have shown that hepcidin regulates iron homeostasis and that various factors regulate its expression [21]. Hepcidin is known to be regulated via the BMP/HJV/SMAD pathway and JAK or IL-6/STAT3 signaling [15, 22, 23]. Moreover, glucose and insulin directly or indirectly upregulate hepcidin [17, 24]. Therefore, iron overload is a common symptom of type 2 diabetes involving insulin secretion disorders [25]. Pgrmc1 regulates hepcidin levels in HepG2 cells and zebrafish [14]. In the present study, we confirmed that Hamp mRNA expression and hepcidin protein levels were decreased in Pgrmc1 KO mice and suggest that Pgrmc1 regulates hepcidin in mammals. Many factors, such as low intercellular and extracellular iron concentrations, inflammation, hypoxia, and erythropoiesis, promote Hamp mRNA transcription. Thus, we investigated the relationship between these factors and Pgrmc1; however, we could not determine the mechanism by which Pgrmc1 is directly regulated in the mouse liver. Hamp mRNA and hepcidin protein levels were significantly different between WT and Pgrmc1 KO mice. This finding suggests that the internal environment of Pgrmc1 KO mice has low hepcidin levels.
Pgrmc1 plays important roles in the body, including insulin secretion, cAMP activation, and hormone synthesis [26, 27]. Pgrmc1 KO mice show slower glucose uptake into the intercellular space than WT mice because of low insulin secretion. This finding suggests that insulin affects the hepcidin levels in Pgrmc1 KO mice. Adenine dietary supplementation in mice leads to increased hepatic hepcidin expression via the BMP/SMAD pathway. Therefore, adenine-mediated cAMP activation increases hepcidin levels [28]. Notably, when cAMP-mediated Creb increases Hamp mRNA transcription [16], Pgrmc1 increases cAMP levels under limited or impaired insulin action [17]. Thus, Pgrmc1 KO mice can potentially have low insulin levels and low cAMP activation. Therefore, these conditions can be used to produce Pgrmc1 KO mice with low hepcidin levels.
Our results described a possible beneficial effect of Pgrmc1 on iron metabolism. Although the direct linking between Pgrmc1 and hepcidin was not recognized in vitro, loss of Pgrmc1 significantly decreased the hepatic hepcidin level. Under the condition of cAMP activation, PGRMC1 induces HAMP mRNA transcription. Considering these results, the internal environment caused by the lack of Pgrmc1 may be a critical factor in anemia.







