Introduction
Recent study showed that the incidence and prevalence of autoimmune disease have been increasing around the world [1]. Autoimmune diseases showed chronic conditions that started the loss of immunologic resistance to the self-antigens [2]. Autoimmune diseases include many diseases, such as vitiligo, celiac disease, and rheumatoid arthritis, autoimmune thyroid disease, psoriasis, adultonset insulin-dependent diabetes mellitus, Addison’s disease, and systemic lupus erythematosus (SLE) [3, 4].
The exact pathogenesis of autoimmune diseases is still unknown. However, recent studies suggested that autoimmune diseases are caused by a combination of individual genetic background and environmental susceptibility factors. Genetic linkage, candidate gene, and advanced genome-wide association studies have implicated a number of autoimmune diseases including vitiligo, celiac disease, and rheumatoid arthritis [5].
Cytokines can play an important role in mediation of immune responses and dysregulation of cytokine production or action. Several studies have suggested that those cytokines have a central role in the susceptibility and development of autoimmune diseases. Among various cytokines, tumor necrosis factor (TNF)-α is a multifunctional and proinflammatory cytokine. Many evidences revealed that TNF-α plays a critical role in the pathogenesis of several autoimmune diseases like autoimmune thytoiditis, rheumatoid arthritis, and diabetes mellitus and several dermatological disorders including vitiligo [6, 7].
The TNF-α gene is located on chromosome 6 (6p 21.31). And its position was included in a major histocompatibility complex (MHC) class III region. There are several single-nucleotide polymorphisms (SNPs) of TNF-α gene. Among these SNPs, specific SNP has been proposed to have the potential to cause structural changes within reg ulatory regions that may affect the function or regulation of TNF-α production [8]. Previous study reported that the promoter SNP at position -308 (-308, G/A) is related to a larger amounts of TNF-α [9] and considered as cause of the dysregulation of the immune system. This has been thought to lead chronic inflammatory processes and autoimmune diseases. Many previous studies tried to clarify the links and meta-analysis was performed to confirm the relationship between vitiligo [10], celiac disease [12], and rheumatoid arthritis [11, 13].
Several studies have attempted to show the relationship between susceptibility to only specific autoimmune disease, such as vitiligo, celiac disease, and rheumatoid arthritis and TNF-α polymorphism (-308, G/A). However, these results are controversial and there is no overall analysis of autoimmune disease. In present study, we performed a meta-analysis using all published data to provide statistical evidence for an association between TNF-α polymorphism (-308, G/A) and autoimmune disease susceptibility.
Materials and Methods
In order to search and include related autoimmune diseases and control studies, PubMed, Google, and Embase database up to October 2016 were investigated. We used various key words: “tumor necrosis factor” or “TNF alpha” or “TNF-α AND polymorphism” or “polymorphisms or variant AND 308” and/or “autoimmune diseases such as vitiligo, celiac disease, and rheumatoid arthritis". The study was restricted to case and control study. Additional studies were found by looking at original articles or review papers as references.
Studies were included if the following criteria were met: (1) evaluated the case and control study between the TNF-α polymorphism (-308 G/A) and autoimmune diseases; (2) used a case-control study design; (3) contained sufficient published data for the estimation of an odds ra- tio (OR) with a 95% confidence interval (CI). Data were extracted from the selected articles including the first author’s name, year of publication, number of autoimmune disease patients and controls and genotype and allele count of TNF-α polymorphism (-308 G/A) in each group.
The correlation between TNF-α polymorphism (-308, G/A) and susceptibility to autoimmune disease was assessed by pooled ORs and corresponding 95% CIs. 95% CI without 1 for OR and p value with <0.05 indicate a significant association with risk of autoimmune diseases. The pooled ORs were calculated for dominant model (G/G genotype vs. G/A+A/A genotypes), recessive model (G/G + G/A genotype vs. A/A genotype) and allelic (G allele vs. A allele). A χ2-test-based Q statistic test was performed to assess heterogeneity of study. I2 test was also performed to assess the effect of heterogeneity. The random-effects Mantel–Haenszel method was adopted if the result of the Q test was p<0.05 or I2 statistic was >50%, which indicated the statistically significant heterogeneity in meta analysis. The fixed effects model was used when there was no significant heterogeneity among the included studies. The statistical analysis was performed by Comprehensive meta analysis (Corporation, NJ, USA). All P values with <0.05 are considered as significant association.
Results
We searched studies about TNF-α polymorphism (-308 G/A) and autoimmune disease in the database including Pubmed, Google, Embase, and Korean database (KISS, RISS, KoreaMed, and KMbase). We finally selected 37 articles for the meta-analysis (Table 1) [10-13]. The 37 articles were including 6,102 patients with autoimmune disease and 6,987 healthy subjects in the current metaanalysis. The data of TNF-α polymorphism (-308, G/A) in the autoimmune disease group and the control group were extracted. These genotype and allele frequencies of TNF-α polymorphism (-308, G/A) were shown in Table 1.
| Authors | Case | Controls | Case | Controls | Diseases | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G/G | G/A | A/A | G/G | G/A | A/A | G | A | G | A | ||
| Naresh et al 2012 | 396 | 436 | 137 | 780 | 184 | 17 | 1228 | 710 | 1744 | 218 | vitiligo |
| Aydingoz et al 2015 | 93 | 12 | 0 | 181 | 27 | 3 | 198 | 12 | 389 | 33 | vitiligo |
| Yazici et al 2006 | 50 | 10 | 1 | 107 | 16 | 0 | 110 | 12 | 230 | 16 | vitiligo |
| Namian et al 2009 | 152 | 17 | 7 | 470 | 73 | 2 | 321 | 31 | 1013 | 77 | vitiligo |
| Al-Harthi et al 2013 | 17 | 103 | 3 | 100 | 76 | 24 | 137 | 109 | 276 | 124 | vitiligo |
| Salinas-Santander et al 2012 | 177 | 21 | 0 | 356 | 36 | 3 | 375 | 21 | 748 | 42 | vitiligo |
| Rossi et al 2015 | 206 | 58 | 3 | 107 | 117 | 20 | 470 | 64 | 331 | 157 | Celiac Disease |
| deAlbuquerque et al 2015 | 42 | 46 | 8 | 100 | 63 | 29 | 130 | 62 | 263 | 121 | Celiac Disease |
| Kekik et al 2011 | 15 | 78 | 0 | 16 | 14 | 3 | 108 | 78 | 46 | 20 | Celiac Disease |
| Capilla et al 2007 | 191 | 60 | 5 | 90 | 44 | 10 | 442 | 70 | 224 | 64 | Celiac Disease |
| Hermann et al 2007 | 148 | 118 | 11 | 8 | 9 | 2 | 414 | 140 | 25 | 13 | Celiac Disease |
| Barisani et al 2006 | 157 | 44 | 1 | 85 | 55 | 15 | 358 | 46 | 225 | 85 | Celiac Disease |
| Garrote et al 2005 | 72 | 26 | 1 | 31 | 18 | 1 | 170 | 28 | 80 | 20 | Celiac Disease |
| Lio et al 2005 | 163 | 53 | 4 | 61 | 34 | 15 | 379 | 61 | 156 | 64 | Celiac Disease |
| Cataldo et al 2003 | 73 | 22 | 1 | 44 | 12 | 12 | 168 | 24 | 100 | 36 | Celiac Disease |
| Hahn et al 2003 | 70 | 27 | 6 | 23 | 44 | 22 | 167 | 39 | 90 | 88 | Celiac Disease |
| Garrote et al 2002 | 51 | 14 | 0 | 20 | 23 | 2 | 116 | 14 | 63 | 27 | Celiac Disease |
| Li et al 2015 | 104 | 8 | 0 | 104 | 23 | 2 | 216 | 8 | 231 | 27 | Rheumatoid arthritis |
| Boechat et al 2013 | 109 | 22 | 0 | 159 | 33 | 0 | 240 | 22 | 351 | 33 | Rheumatoid arthritis |
| You et al 2013 | 422 | 30 | 0 | 323 | 50 | 0 | 874 | 30 | 696 | 50 | Rheumatoid arthritis |
| Al-Rayes et al 2011 | 68 | 35 | 3 | 63 | 48 | 15 | 171 | 41 | 174 | 78 | Rheumatoid arthritis |
| Hussein et al 2011 | 134 | 36 | 2 | 150 | 10 | 0 | 304 | 40 | 310 | 10 | Rheumatoid arthritis |
| Emonts et al 2011 | 248 | 119 | 8 | 300 | 147 | 14 | 615 | 135 | 747 | 175 | Rheumatoid arthritis |
| Trajkov et al 2009 | 67 | 15 | 2 | 231 | 66 | 4 | 149 | 19 | 528 | 74 | Rheumatoid arthritis |
| Ates et al 2008 | 73 | 25 | 0 | 101 | 21 | 0 | 171 | 25 | 223 | 21 | Rheumatoid arthritis |
| Rezaieyazdi et al 2007 | 29 | 5 | 0 | 29 | 1 | 0 | 63 | 5 | 59 | 1 | Rheumatoid arthritis |
| Nemec et al 2008 | 93 | 36 | 1 | 121 | 29 | 0 | 222 | 38 | 271 | 29 | Rheumatoid arthritis |
| Rodriguez et al 2005 | 113 | 17 | 3 | 148 | 14 | 0 | 243 | 23 | 310 | 14 | Rheumatoid arthritis |
| Correa et al 2005 | 109 | 52 | 4 | 338 | 87 | 5 | 270 | 60 | 763 | 97 | Rheumatoid arthritis |
| Pawlik et al 2005 | 74 | 17 | 0 | 77 | 25 | 3 | 165 | 17 | 179 | 31 | Rheumatoid arthritis |
| Balog et al 2004 | 14 | 9 | 0 | 51 | 22 | 2 | 37 | 9 | 124 | 26 | Rheumatoid arthritis |
| Cunenca et al 2003 | 71 | 20 | 1 | 38 | 4 | 0 | 162 | 22 | 80 | 4 | Rheumatoid arthritis |
| Yen et al 2001 | 94 | 3 | 0 | 72 | 23 | 2 | 191 | 3 | 167 | 27 | Rheumatoid arthritis |
| Van et al 1999 | 195 | 76 | 12 | 79 | 52 | 7 | 466 | 100 | 210 | 66 | Rheumatoid arthritis |
| Vinasco et al 1997 | 43 | 14 | 3 | 84 | 16 | 2 | 100 | 20 | 184 | 20 | Rheumatoid arthritis |
| Danis et al 1995 | 17 | 13 | 4 | 44 | 13 | 0 | 47 | 21 | 101 | 13 | Rheumatoid arthritis |
| Fugger et al 1989 | 15 | 6 | 3 | 63 | 60 | 8 | 36 | 12 | 186 | 76 | Rheumatoid arthritis |
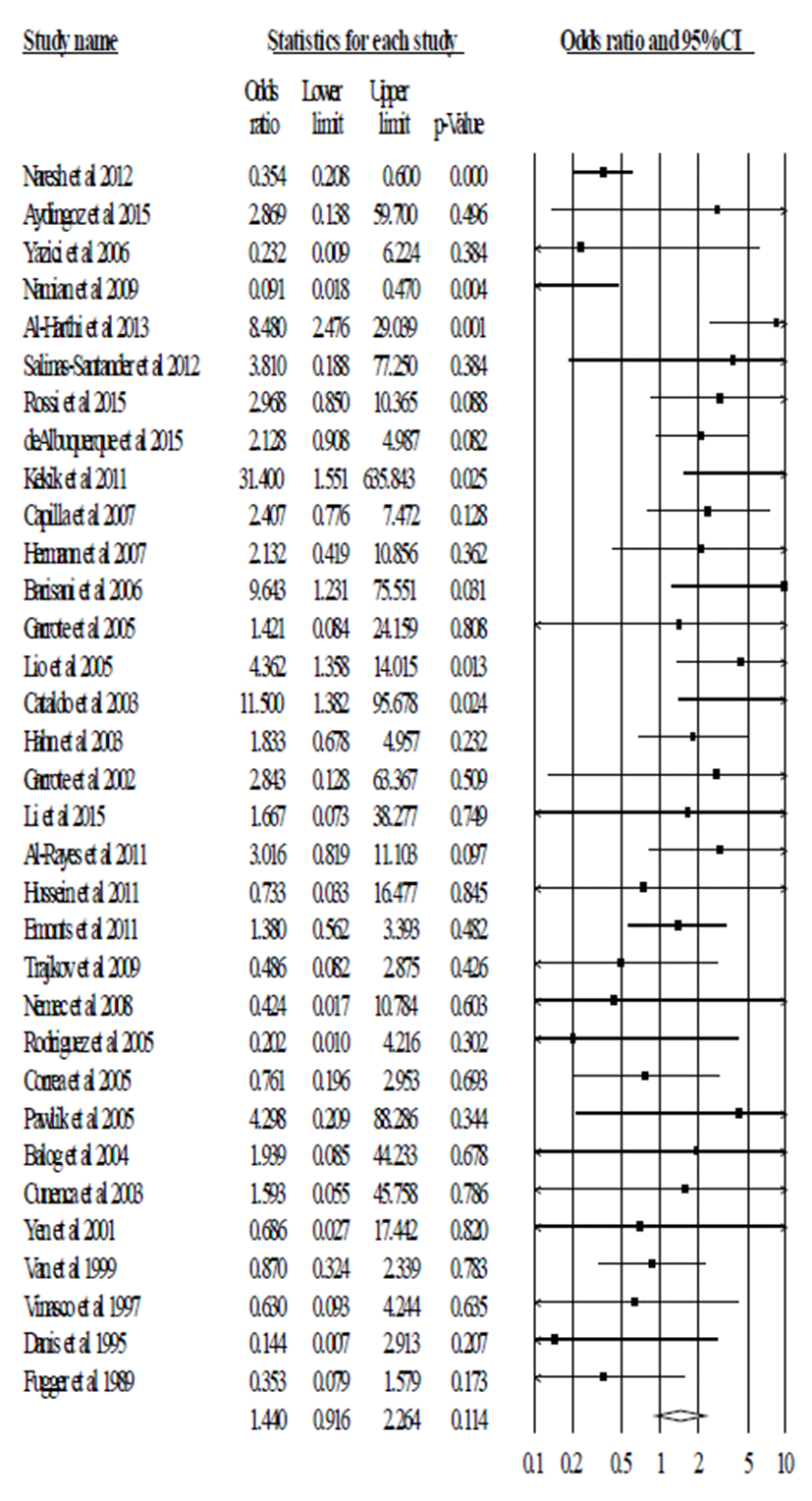
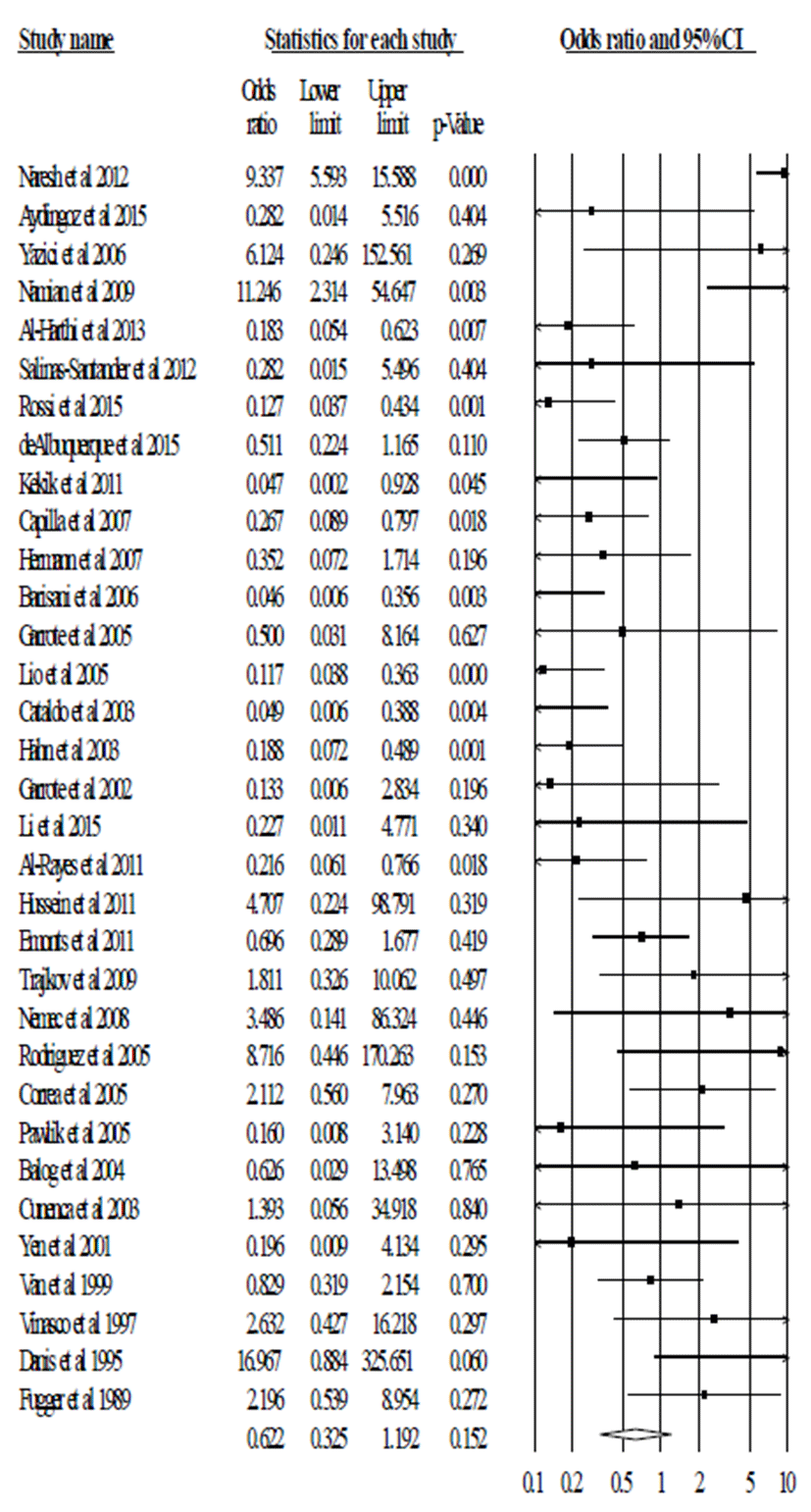
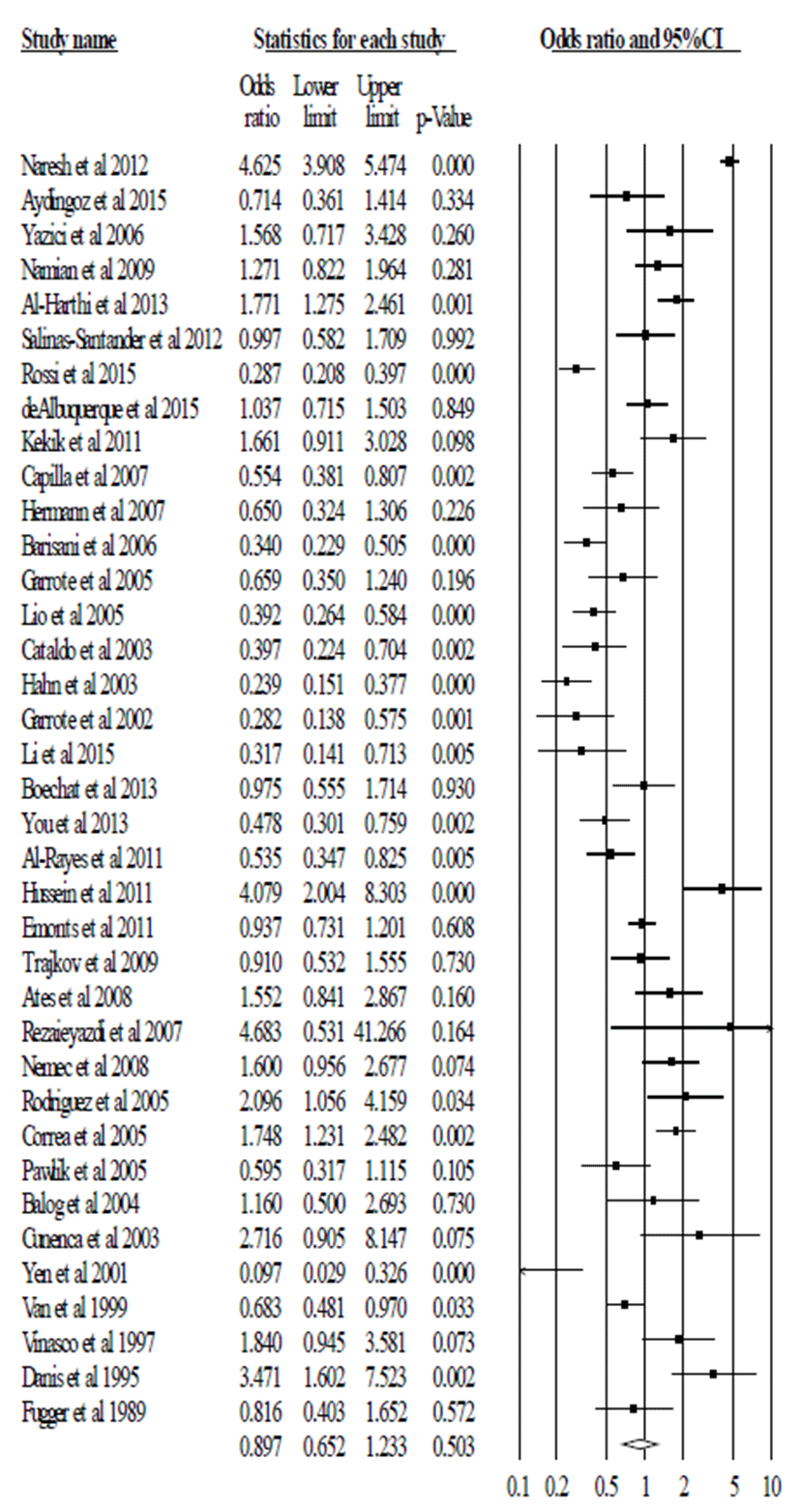
In total, 6,102 patients with autoimmune disease and 6,987 healthy subjects from 37 articles were analyzed for the correlation between TNF-α polymorphism (-308, G/A) and susceptibility to autoimmune diseases. We compared the frequencies of genotype combination and allele frequency. The pooled ORs were calculated for dominant model (G/G vs. G/A+A/A), recessive model (G/G+G/A vs A/A) and allelic model (G allele vs. A allele). We firstly calculated the p value and I2 for heterogeneity. These values were <0.05 and <60.00. So, we applied the random model.
In pooled analysis, we did not observe the significant association between TNF-α polymorphism (-308, G/A) and risk of autoimmune disease in recessive model (G/G+G/A vs A/A, OR=0.622, 95% CI=0.325-1.192, p=0.152 in Fig. 1 and Table 2), dominant model (G/G vs. G/A+A/A, OR=1.440, 95% CI=0.916-2.264, p=0.114 in Fig. 2 and Table 2), and allele model (G vs. A, OR=0.897, 95% CI=0.652-1.233, p=0.503 in Fig. 3 and Table 2). These results indicate that TNF-α polymorphism (-308, G/A) may not be associated with susceptibility to autoimmune diseases.
Discussion
TNF-α is a potent proinflammatory cytokine that acts in the T helper 1 immune response. In vitiligo disease, TNF-α is associated with T-cell trafficking to the skin and T-cell/melanocyte attachment, augmenting destruction of melanocytes [14]. In addition, TNF-α affects the apoptotic pathway of melanocytes [15]. Many studies have observed various TNF-α production in the skin, peripheral blood and serum of vitiligo patients.
In case-control study, it is suggested that the minor allele TNF-α (-308, G/A) affected increased plasma level of TNF-α with an increased risk for various disease, including rheumatoid arthritis [16], Hashimoto thyroiditis [17], diabetes mellitus [18], etc. Also, in meta-analysis study, TNF-α gene polymorphisms have been investigated in the association with susceptibility to rheumatoid arthritis [11], insulin resistance [19], autoimmune hepatitis [20], sarcoidosis [21], Alzheimer’s disease [22], asthma [23], and cancers [24].
To explore the potential relationship of TNF-α (-308, G/A) polymorphism with each autoimmune disease risk, several case-control studies have been investigated. These results were controversial. Al-Harthi et al. proposed a direct association between TNF-α (-308, G/A) polymorphism and vitiligo in Saudi patients [25]. The study reported that TNF-α (-308, G/A) polymorphism in vitiligo patients showed higher frequency of GA genotype and lower frequency of G/G and A/A genotype compared to that in controls. Laddhaet al. found that TNF-α (-308, G/A) allele increased the risk of generalized vitiligo by 4.326 fold in India patients [26]. Namian et al. reported similar association between TNF-α (-308, G/A) polymorphism and vitiligo in Iranian patients [27]. Salinas- Santander et al. suggested a possible association between G/A and A/A genotype and the active form of vitiligo in Mexican population [28]. However, Yaziciet al. observed a lack of association between TNF-α (-308, G/A) polymorphism and vitiligo among Turkish patients [29].
In the current meta-analysis, we addressed the relationship between TNF-α polymorphism (-308 G/A) and susceptibility to overall autoimmune disease. In the present meta-analysis, a total of 6,102 patients with autoimmune disease and 6,987 healthy subjects from 37 case–control studies were included. Meta-analysis of TNF-α polymor- phism (-308, G/A) did not detect any significant association with autoimmune diseases.
There are several limitations in the current meta-analysis. First, the etiological mechanism of autoimmune diseases is very complicated, in which gene-gene, and gene-environment interactions are involved. Association with autoimmune diseases could not easily be detected by the meta-analysis. Second, the numbers of studies and individual sample sizes included in our pooled analysis were not sufficiently large for a comprehensive analysis, especially for ethnicity-based subgroup analyses, and thus there is room for further study.
Our results demonstrate that TNF-α (-308, G/A) polymorphism may not be contributed to autoimmune disease susceptibility. However, It is necessary to study these autoimmune diseases in more races and to confirm these results at sufficient sample size.







