Introduction
Discovery of the association of Helicobacter pylori with gastritis, peptic ulcers, and gastric neoplasia has led to fundamental changes in the understanding of gastric disease in humans [1, 2, 3]. Like H. pylori, other species of Helicobacter have been shown to colonize the stomach and cause disease in animals. Gastric colonizers include H. felis, H. mustelae, H. acinonychis, H. bizzozeronii, H. heilmannii, H. salomonis, and a recently isolated novel Helicobacter sp. in dolphins [4, 3]. H. felis is a gram-negative, spiral-shaped bacterium originally isolated from the stomachs of cats and dogs [5]. This organism has been shown by 16S rRNA gene sequencing to be very closely related to H. pylori [6]. Further, H. felis has been shown to colonize mice and elicit host reactions and histopathology, which are very similar to those seen in H. pylori infections of humans [5].
In human, H. pylori is a major Helicobacter species. H. pylori produce large amounts of urease in order to catalyze urea hydrolysis. Urease neutralizes stomach acid by generating ammonia from urea, which is essential for survival of H. pylori in the host [7, 8]. Thus, H. pylori diagnosis is based on detection of urease. Several methods have been proposed and used to diagnose H. pylori infection. Increasing interest has been directed toward non-invasive tests compared to endoscopy-based invasive methods (histology and rapid urease test), as non-invasive methods do not require endoscopic assessment [9]. The 13C-urea breath test (UBT) is the most recommended non-invasive test for detection of H. pylori infection and has high sensitivity and specificity [10]. However, the UBT cannot be applied to animals due to its high cost and requirement for expensive analytical instruments [11]. Thus, many researchers have used polymerase chain reaction (PCR) assays to monitor H. pylori infection in stools since it does not require biopsy nor animal sacrifice [12, 13]. However, PCR assay is time-consuming and involves high-cost laboratory instruments like a thermal cycler [1]. Furthermore, stool samples remain the most difficult specimens for DNA extraction and amplification [14]. Recently, several companies have released H. pylori stool antigen (HpSA) test kits, which are non-invasive diagnostic modules for detecting H. pylori infection using human patient’s stool samples [15, 16]. However, there is little information about the usefulness of HpSA tests for H. felis, which is a major Helicobacter species in dogs and cats.
The aim of the present study was to compare diagnostic methods for diagnosis of H. felis using HpSA tests and PCR assay using cat stools or gastric mucosa.
Materials and Methods
H. felis (ATCC 49179; American Type Culture Collection, USA) was incubated in brain-heart infusion broth containing 10% fetal bovine serum at 37°C overnight under a microaerophilic atmosphere and allowed to grow to a density of ~2.0 × 109 colony-forming units (CFU) per 1 mL of culture broth.
Six male 8-month-old healthy cats were obtained from the animal facility of Wonkwang University and housed in a room with constant environmental conditions (22 ± 2°C; 40~70% relative humidity; 12-hour light-dark cycle; 150~300 lux brightness). Pellet feed and purified water were available ad libitum. All animal experiments were conducted according to Standard Operation Procedures and were approved by the Institutional Animal Care and Use Committee of Wonkwang University, Korea.
Six male cats were used. After a 24-hour fast, cats were orally inoculated twice at 3-day intervals by oral administration of 1.0 × 109 CFU of H. felis suspended in 10 mL of broth.
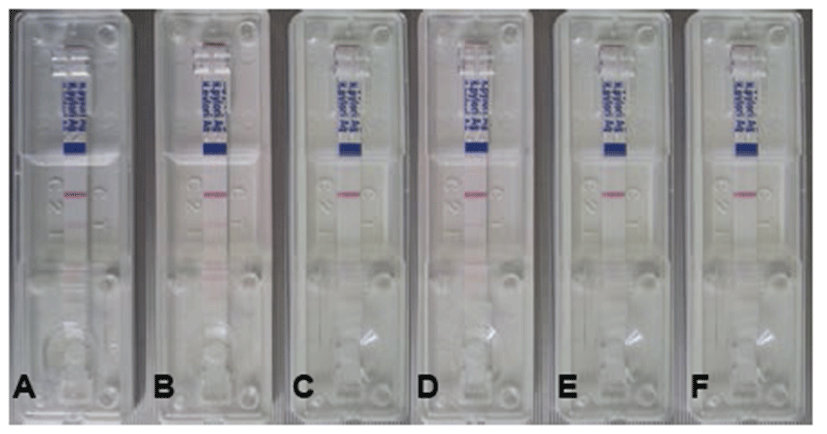
After H. felis inoculation, stool specimens were gathered on days 1, 3, 5, 7, 14, and 21. The H. felis antigen was evaluated using a commercially available SD Bioline H. pylori Ag kit according to the manufacturer’s instructions. Specimens (250 mg) were incubated with diluent solution at room temperature for 30 min, after which 100 μL was placed on the Helicobacter Ag examination device. The test results were checked about 15 min later. One red line indicates a negative result, and a double red line indicates a positive Helicobacter result.
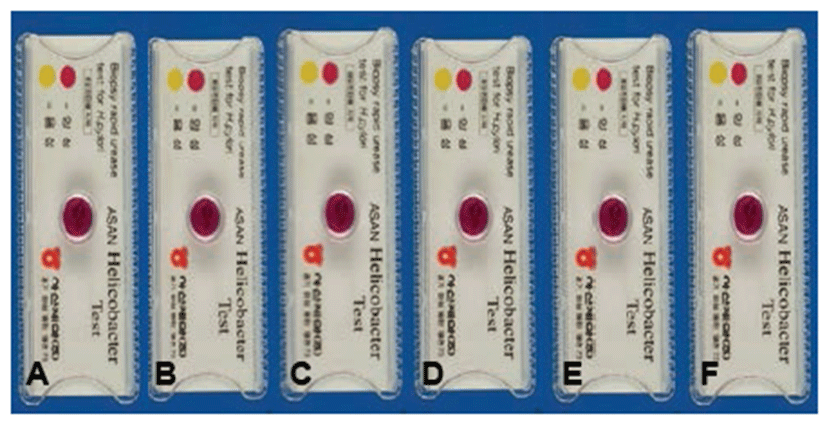
The cats were sacrificed 21 days after H. felis inoculation, and their gastric mucosa was biopsied to detect H. felis. To confirm infection using a rapid urease test (RUT), biopsy samples (3 × 3 cm) from the gastric pylorus were minced and applied to CLO Helicobacter-detection kits (Asan Pharm Co., Ltd., Seoul, Korea) by incubating biopsy specimens at 35°C for 24 hours to examine urease activity. The reaction (color change) was classified as bright yellow (negative), thick yellow false (partially) positive, or thick (dark) red for positive.
After inoculation, stool specimens were gathered on days 1, 3, 5, 7, 14, and 21. Stool samples of H. felis-infected cats were homogenized and resuspended in PBS, and DNAs were extracted using the AccuPrep Stool DNA Extraction Kit (Bioneer, Korea) according to the manufacturer's instructions. AccuPrep Stool DNA Extraction Kit is designed for the rapid, convenient extraction of DNA from fresh or frozen stool, or other samples containing large amounts of material that can inhibit PCR. The kit uses a glass filter fixed in a column tube that can efficiently bind DNA in the presence of chaotropic salts. Using the spin-column method, contaminants and enzyme inhibitors (such as heparin, bilirubin bile salts, and porphyrin) are eliminated, and high-purity DNA is obtained that is ready for use in a variety of applications [2]. After washing to remove proteins and salt, high-purity DNA is finally eluted using a low-concentration elution buffer. The kit provides 2~5 μg of DNA from 100 mg of stool [2].
The cats were sacrificed 21 days after H. felis inoculation, and their gastric tissues were homogenized and resuspended in PBS. DNAs were extracted from the homogenates of gastric tissues after their isolation using an AccuPrep Genomic DNA Extraction kit (Bioneer Corp., Daejeon, Korea), according to the manufacturer’s instructions.
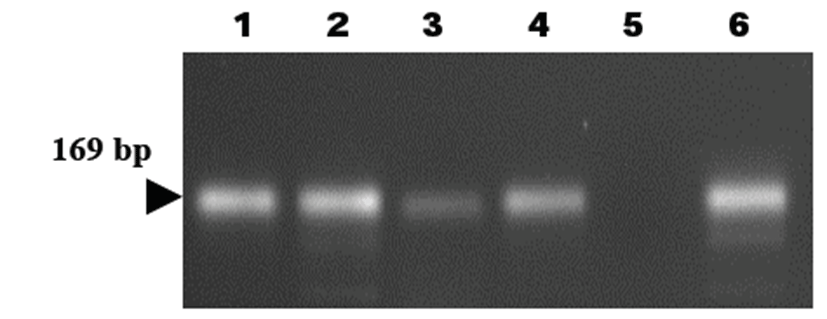
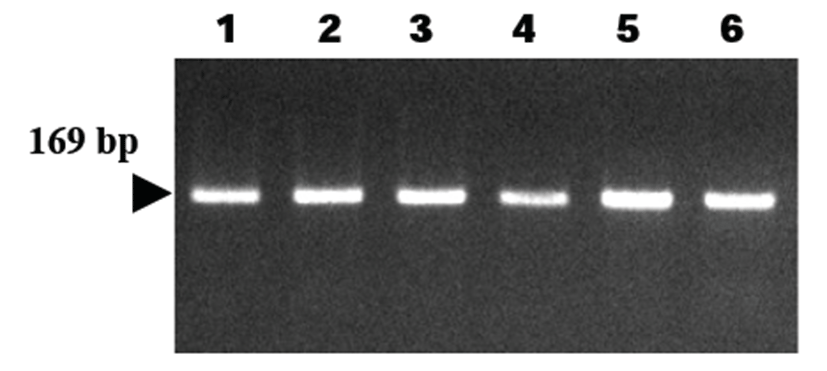
DNAs were eluted in Tris-EDTA buffer (pH 8.0), and an aliquot was used for PCR amplification. All DNA samples were stored at – 20°C until PCR assays were performed. Primers have been designed to amplify the 16S rRNA gene of H. felis [17]. The sequence of the H. felis primer HF-L is 5’-ATGACATGCCCTTTAGTTTGGGATAGCCA-3’, whereas that of primer HF-R is 5’-CGTTCACCCTCTCAGGCCGGATACC-3’. This primer set is known to amplify a 169 bp fragment and has been used for detection of H. felis in samples (Kong et al., 1996).
The template DNA (400 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer) containing 2.5 U of Taq DNA polymerase, 250 μM of each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and gel loading dye. The final volume was adjusted to 20 μL with distilled water. The reaction mixture was subjected to denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 20 sec, 55°C for 30 sec, 72°C for 1 min, and a final extension step of 72°C for 10 min. Samples were maintained at 4°C until analysis. Reactions were conducted using My Genie 32 Thermal Block PCR (Bioneer). Eight microliters of each sample was mixed with 2 μL of loading buffer and electrophoretically separated on 2.0% agarose gels stained with 0.5 μg/mL of ethidium bromide. DNA bands were observed under ultraviolet light.
Statistical analysis was performed using analysis of variance and SPSS for Windows v.12.0 (Chicago, IL, USA). A P-value<0.05 was considered statistically significant. The positive ratio was measured using MINITAB software and 95% confidential intervals (Minitab Inc., State College, PA, USA). A positive ratio indicated a significant difference.
Results
After final inoculation of H. felis into cats, we collected stool specimens on days 1, 3, 5, 7, 14, and 21. As the stools contained low amounts of water, we incubated the stool specimens in diluent solution for 30 min with vortexing every 10 min. Then, we placed 100 μL of sample on the SD Bioline H. pylori Stool Ag kit detection device.
On day 1, we observed no positive results. However, the positive ratio increased on days 3 [16.7% (1/6)], days 5 [16.7% (1/6)], and days 7~21 [50.0% (3/6)] (Table 1). One red line indicates a negative result, and a double red line indicates a positive Helicobacter result (Fig. 1).
PCR analysis based on the 16S rRNA gene of H. felis was employed to detect H. felis infection. The 16S rRNA gene of H. felis was specifically amplified by PCR using 16S rRNA gene-specific primers (HF-L and HF-R). Five of six stool samples were positive in the PCR reaction (5/6, 83.3%) (Table 2). The target nucleic acid fragments were specifically amplified by PCR using the 16S rRNA primers (Fig. 2).
Repeated intragastric inoculation of H. felis into cats resulted in positive reactions (red color) 21 days after the final inoculation, indicating bacterial infection in the stomach (Fig. 3). All gastric biopsy specimens were positive in the RUT on day 21 (6/6, 100%) (Table 2).
Cats were sacrificed 21 days after H. felis inoculation, and their gastric tissues were homogenized and resuspended in PBS. DNAs were extracted from the homogenates of gastric tissues, and PCR analysis was employed to detect H. felis infection. As a result, all samples were positive in the PCR reaction (6/6, 100%) (Table 2, Fig. 4).
Discussion
Diagnosis of H. pylori infection can be made using both invasive and non-invasive tests. Invasive tests include histology, culture, and the rapid urease test (RUT), which requires endoscopy to obtain gastric mucosa biopsies. Non-invasive tests for diagnosis of H. pylori infection based on analysis of breath, blood, and stool samples have been developed [18]. However, serological tests are unable to distinguish active from past infections [19]. Non-invasive tests require that the microorganism be detected in gastric biopsy samples, which means an endoscopy must be performed. The current gold standard for diagnosis of H. pylori infection is endoscopic biopsy of gastric tissue for RUT, histology, and culture. However, such an invasive procedure has major disadvantages, including anesthesia, discomfort, and possible ethical problems [20]. On the other hand, non-invasive tests are easy to perform and do not produce significant discomfort nor risk of invasive endoscopy. These tests include serological antibody testing for H. pylori, urea breath test (UBT), and stool antigen assay (HpSA) test [20].
Among these several diagnostic tests, the HpSA test and PCR using stool specimens for diagnosis of H. felis infection are non-invasive methods that do not require animal sacrifice in an in vivo study or veterinary clinic. In this study, sensitivities of the HpSA test and PCR analysis using stool specimens were 50.0% and 83.3% respectively. On the other hand, sensitivities of the RUT and PCR using gastric specimens were 100% and 100% respectively. Despite the good performance of these H. felis antigen detection tests using stool samples, one disadvantage is that antigen excretion may vary over time and antigens may degrade while passing through the intestine. Further, use of a mucolytic agent like N-acetylcysteine may decrease accuracy of the diagnosis [21]. The cut-off titer in antigen detection is difficult to determine but crucial. Stool antigen detection using monoclonal antibody is recommended by the European Helicobacter Study Group since it provides equivalent diagnosis accuracy as the UBT [22]. In this study, RUT and PCR using gastric specimens were highly sensitive for detection of H. felis infection, although they cannot be applied to live animals since sacrifice is required.
In this study, PCR analysis was more sensitive than the HpSA test using stool specimens. Since the HpSA test is designed to target H. pylori antigen, it may not be able to completely detect whole H. felis antigen. Therefore, for non-invasive diagnosis of H. felis infection using animal stool samples, PCR analysis is more sensitive. However, PCR analysis has several reported limitations [1]. There is no single gold standard among diagnostic tests for detection of Helicobacter infection, but all tests have their pitfalls and limitations [23]. Among non-invasive methods, stool specimens are easy to obtain and consequently have high potential for the development of a method for direct detection of Helicobacter species [1, 2]. PCR was successfully used to detect this bacterium in stools [1, 2]. Indeed, stool samples remain the most difficult specimens for DNA extraction and amplification [14] due to the presence of DNA polymerase inhibitors identified as complex polysaccharides [14]. In this study, we used the spin-column method to effectively eliminate contaminants and enzyme inhibitors (such as heparin, bilirubin bile salts, and porphyrin) and obtain 2~5 μg of high-purity DNA from 100 mg of stool [24]. Although the HpSA test was shown to be less sensitive, it could be used for rapid, cheap, and easy screening of H. felis infection in dogs and cats based on positive results of H. felis-specific PCR.
We suggest that the H. pylori stool antigen kit, HpSA, is useful and effective for monitoring H. felis infection in stool specimens. If the HpSA test could be made with H. felis antibodies in the future, its sensitivity could be increased. Further, PCR assay could be successfully used to detect H. felis in stools. Application of the HpSA kit and PCR assay can be an effective non-invasive strategy to identify H. felis in cats.







