Introduction
The immune system in humans is classified into two krsubsystems, innate versus adaptive immune and humoral versus cell-mediated immunity. The immune system constantly interacts with pathogenic and non-pathogenic bacteria, including Gram-negative/positive bacteria [1]. Although commensal strains such as lactic acid bacteria and bifidobacteria are tolerated in the gut, pathogenic strains are restrained by the immune system. However, neither commensals nor pathogenic bacteria are tolerated by the immune system outside of the gastrointestinal track. Commensal bacteria are reported to inhibit growth of pathogenic bacteria through acidic metabolites and subsequent creation of an acidic gut environment [2].
Bacterial infection across the intestinal surface barrier into the underlying gut tissue stimulates activation of nearby immune cells by bacterial antigens such as peptidoglycan, outer membrane protein A (OmpA), flagella, and bacterial DNA [3-5]. Lipopolysaccharide (LPS)/OmpA and muramyl dipeptide (MDP) are structural components of pathogenic bacteria that act as potent antigenic activators of the immune system as well as downstream signal toll like receptor (TLR) pathways [6-9]. On the other hand, short chain fatty acids (SCFAs) are known to be the most abundant microbial metabolites in the intestine, leading to activation of MAPK signaling and rapid production of chemokines and cytokines [10]. These pathways mediate protective immunity and tissue inflammation in mice [11]. Acetic or butyric acids are known to induce unique intracellular pH changes [12] or anti-inflammatory activity [13].
Macrophages, a type of white blood cells found in all tissues, engulf and digest cellular debris, foreign substances, microbes, and cancer cells in a process called phagocytosis [14]. Macrophages play a critical role in non-specific defense (innate immunity) and help initiate specific defense mechanisms (adaptive immunity) by recruiting other immune cells such as lymphocytes. In addition, they play an important anti-inflammatory role and can reduce immune reactions through release of cytokines. However, most of these studies were performed in human lymphocytes, including macrophages [15].
There is a need to identify novel biomarkers of the immune response in domestic animals. In the present study, we attempted to characterize the gene expression profile of porcine macrophages treated with LPS/OmpA/MDP or acetic/butyric acid by DNA microarray and RT-PCR assay.
Materials and Methods
Monocytes/macrophages from pig blood (PoM2) (Pig Monocytes clone 2) were established at the University of Maribor Slovenia (Gradišnik et al., 2006). They were grown in advanced Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich, Grand Island, USA), supplemented with 5% fetal calf serum (Lonza, Basel, Switzerland), L-glutamine (2 mM, Sigma), penicillin (100 U/mL, Sigma), and streptomycin (1 mg/mL, Fluka, Buchs, Switzerland).
LPS and MDP were purchased from Sigma Chemical Co. (Mo., USA). OmpA antigen was kindly provided by Professor Kang, Ho Young at Pusan National University.
AffymetrixGeneChip® Porcine Genome Array was used to analyze the gene expression profile of the porcine macrophage cell line-Pom2. After stimulation with bacterial antigen such as LPS, OmpA, or MDP for 6 hrs, total RNAs were extracted with an RNA Blood Mini Kit (QIAGEN, USA) for analysis of differentially expressed genes by an AffymetrixGeneChip® Scanner 3000 7G, Affymetrix Command Console1.1 and MAS5 Normalization (Macrogen, Seoul, Korea).
POM2 cells were seeded in a 35-mm dish at a density of 2 × 105 cells/well overnight, followed by treatment with PBS, MDP (1 μg/mL), LPS (1 μg/mL), or OmpA (5 μg/mL) for 6 hrs. Total RNAs were extracted with an RNA Blood Mini Kit (QIAGEN, USA) before cDNA synthesis with AccuPower rocket script RT/PCR premix (Bioneer) and 10 ng of total RNA and 10 μM of 5’ and 3’ primers (each) in a total volume of 20 μL. PCR reaction was as follows: cDNA synthesis for 42ºC/40 min and pre-denaturation for 95ºC/5 min, followed by 35 cycles of denaturation for 95ºC/30 sec, annealing for 54ºC/30 sec, and extension for 72ºC/30 sec. PCR reaction (10 μL) was run on 1.5% agarose gels for electrophoresis and stained with ethidium bromide for image analysis.
Results
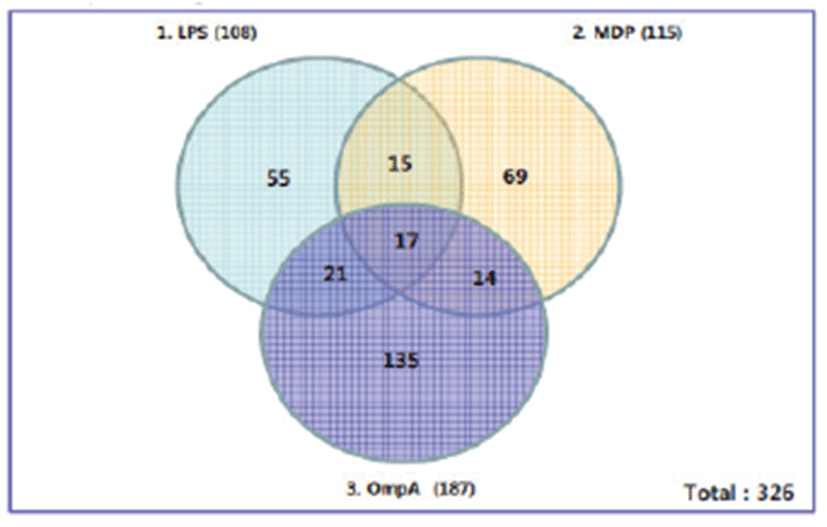
Gene expression assay using immune cells is a common way to investigate the diverse roles of macrophages in bacterial infections or antigenic challenges. Thus, we chose DNA microarray to chart the expression levels of porcine genes in response to bacterial antigens such as LPS, MDP, and OmpA. Pom2 macrophage cells were treated for 6 hrs with each antigen, after which total RNAs were extracted for GeneChip® Porcine Genome Array analysis. Of about 20,000 genes in the array, a total of novel 326 genes were found to be differentially expressed by at least 2-fold upon antigen stimulation compared to the control. Among these, LPS differentially regulated the expression of total 108 genes; 86 genes were up-regulated and 22 genes were down-regulated (Fig. 1). The list in Table 1 shows 16 novel genes representing diverse biological processes that were well annotated and up-regulated by at least 2-fold upon LPS stimulation. Pappalysin-2 is a metalloproteinase involved in cartilage development and angiogenesis without any known role in the immune response. SLC37A2 (Sugar phosphate exchanger) is a novel gene involved in glucose 6 phosphate transport but without any apparent role in the immune response. IL-23 along with IL-17 may participate in an acute response to infection in peripheral tissues, whereas CD1d is known to mediate presentation of primarily lipid and glycolipid antigens of self or microbial origin to T cells. Expression levels of the 16 genes above were also up-regulated by 2-folds upon MDP and OmpA stimulation
Based on the list of up-regulated porcine genes, we confirmed the DNA microarray data by RT-PCR. Using the gene-specific primers shown in Table 2, 10 novel porcine genes along with one control GAPDH gene were chosen for RT-PCR analysis (Fig. 2). Pom2 macrophage cells were stimulated with each of the three antigens above before total RNA extraction. Out of the 10 candidate genes for bacterial antigen responsive biomarkers, SLC37A2, PAPPA2, TMEM 184C, and Syntaxin-binding protein 5 (Stx5) were shown to be up-regulated by RT-PCR. Although the expression level of porcine Stx5 was modestly up-regulated in the DNA microarray analysis (1.2-fold), expression of its human homolog increased by more than 4-fold in human macrophage THP1 cells compared to the negative control (results from another DNA microarray experiment and data not shown). Further, its expression level was up-regulated by an average of 5-fold in response to each antigen, as shown in Fig. 2.
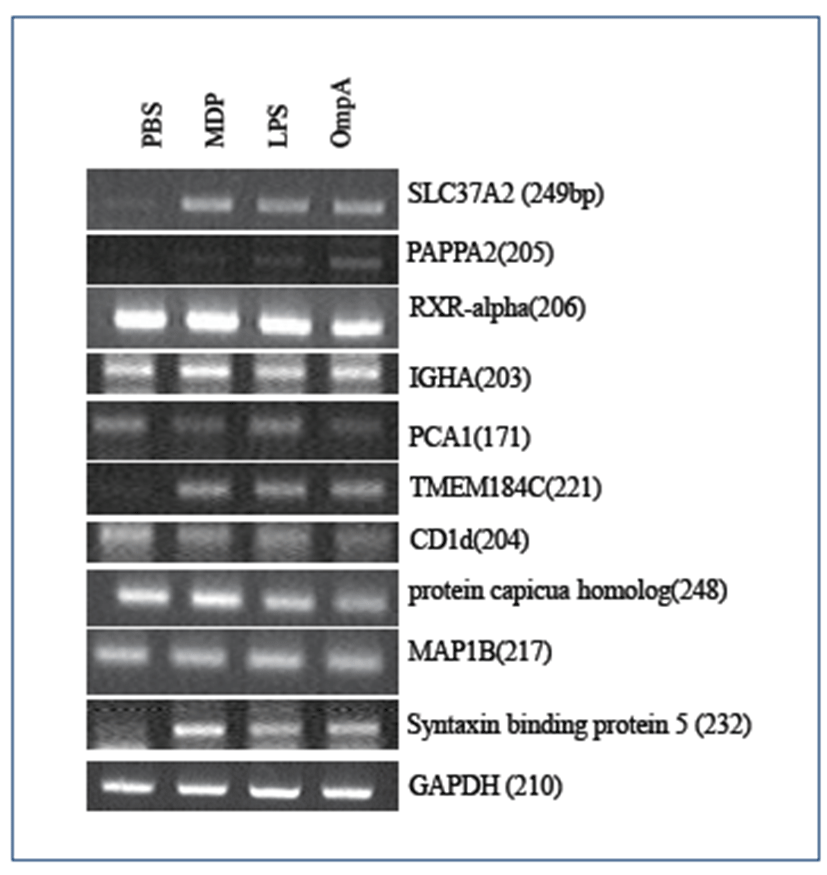
Regardless of the class of each antigen (lipopolysaccharide vs proteinaceous), the three antigens had similar effects on the expression of the four genes (Fig. 2). However, it is unclear why the RXR-alpha, IGHA, PCA1, CD1d, protein capicua homolog, and MAP1B genes did not show any differential expression patterns compared to the control.
SCFAs are abundant in the intestines of vertebrates and are proposed to play a role in the immune response in addition to their direct anti-microbial activity. Therefore, we investigated the effects of two major SCFAs, acetic and butyric acid, on expression of major porcine antimicrobial peptide (AMP) genes. Concentrations of acids were determined based on the minimal amount of each acid that lowered the pH of the culture media (DMEM with 10% FBS) and induced a color change by the pH indicator phenol red. The pH levels of the culture media containing 0.5, 2, and 5 mM of each acid were 8.41, 8.22, and 7.86 for acetic acid as well as 8.12, 8.04, and 7.75 for butyric acid, respectively, whereas the pH of control DMEM with 10% FBS was 8.37.
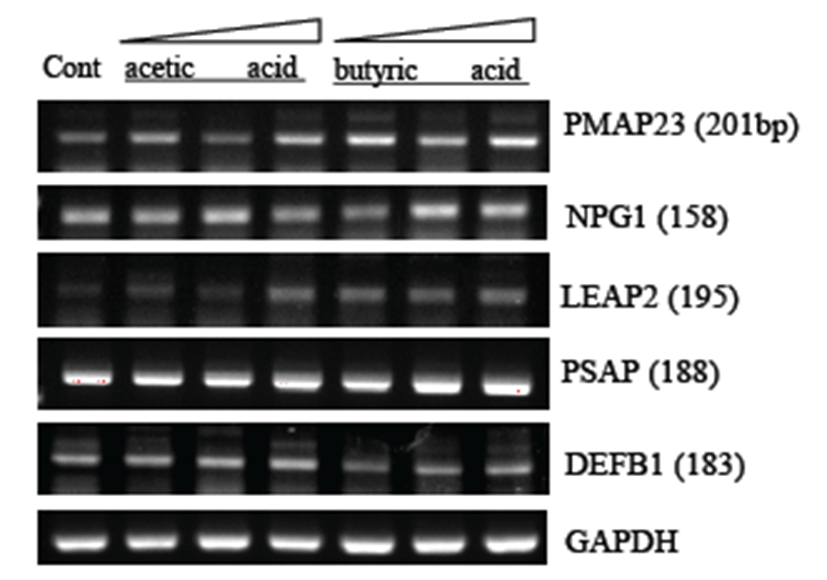
The porcine genome has been sequenced, but its annotation is incomplete. Professor Kim, Herbal at Seoul National Univ. helped us find human homologs of the antimicrobial genes listed in Table 3. Pom2 cells were treated with the acids at final concentrations of 0.5, 2, and 5mM for 15 hrs before total RNA extraction and RT-PCR analysis using gene-specific primers (Table 3). Out of seven porcine AMP genes, including PMAP-23, NPG1, LEAP2, NPG3, BPIFA1, PSAP, and DEFB1, expression of three genes (PMAP23, NPG1, and LEAP2) was up-regulated by butyric and acetic acids. Butyric acid appeared to have better activity than acetic acid in this experiment.
Discussion
Identification of biomarkers is regarded as an important step towards disease characterization and management of disease in animals. Post-genomic technologies have promoted the development of strategies aimed at identifying specific and sensitive biomarkers from the thousands of molecules present in tissues or biological fluids [16].
In this study, we searched for porcine biomarkers showing induced expression in response to three bacterial antigens (LPS, MDP, and OmpA) as well as bacterial metabolites of short chain fatty acids (acetic and butyric acid).
Porcine DNA microarray analysis of Pom2 cells stimulated by these three antigens revealed a total of 326 porcine genes showing at least 2-fold increased expression (Fig. 1). Expression of 16 genes increased by 2-fold regardless of the type of antigen; LPS and MDP are components of peptidoglycan, whereas OmpA is the major protein in the outer membrane region. We also examined half of the 16 representative genes by RT-PCR. Out of 10 porcine genes, four genes (SLC37A2, PAPPA2, TMEM184C, and Stx5) showed differential expression (Table 2 and Fig. 2). Although we used the same RNA samples for DNA microarray and RT-PCR experiments, it is unclear why the other six genes did not show differential expression. There might have been differences in the probing region of each transcript by either the probes of the DNA microarray or RT-PCR primers. Splicing variations might also have caused these differences. Recently, the expression of 22 inducible genes, which include VCAM1, HMOX1, and Serglycin, was reported to be induced in response to Salmonella or LPS in porcine alveolar macrophages. Interestingly, 13 genes, including IL1-a/-b, CD14, OPN, and VCAM, were found to be putative NF-kappaB targets [17]. However, there is no report on the role of NFkB in the transcriptional regulation of the four genes identified in this study (SLC37A2, PAPPA2, TMEM184C, and Stx5).
SCFAs have been reported to be major intestinal microbial metabolites and include acetic/butyric acids. These appear to be absorbed into the body either through specific receptors or passively. Most SCFA studies have focused on their direct inhibitory effects on pathogenic microbes [18], but recent studies have investigated their effects on the host metabolism, including gluconeogenesis, insulin secretion, and fatty acid metabolism [19-21].
Until now, there is no report that describes their effect on innate immunity. Therefore, we studied the effects of SCFAs on expression of a set of antimicrobial peptide (AMP) genes in Pom2 porcine macrophage cells (Table 3). Out of five porcine homologs of human AMP genes investigated by RT-PCR, three genes (PMAP23, NPG1, and LEAP2) were found to be up-regulated by both acetic and butyric acids (Fig. 3). It remains to be characterized which signaling pathway is involved in the modulation of AMP expression and the innate immune response against any invading microbe. It will be interesting to compare the innate immune response between human and porcine macrophages for either antigens or bacterial pathogens. In conclusion, we report here a set of novel porcine biomarkers involved in the innate immune response of porcine macrophages. This finding may lead us to better understand the porcine immune response and develop diagnostic biomarkers for the prevention or diagnosis of pathogenic bacteria such as Salmonella typhimurium.