INTRODUCTION
Histopathological diagnosis procedure involves collecting animal tissues through necropsy, followed by fixation, trimming, tissue processing, embedding, and sectioning. The thin tissue sections obtained through these processes are placed on glass slides and stained [1]. The stained tissue slides are observed under a microscope to provide pathological information. Toxicopathologists may need to examine between 100 and 10,000 slides, which is highly time-consuming and labor-intensive, leading to increased fatigue and potentially affecting diagnostic accuracy. Furthermore, diagnoses made by the pathologists often rely on subjective judgment. To ensure accuracy, a peer review process involving consultations with other pathologists is necessary to finalize the diagnosis. However, during the process of transferring slides for peer review, there is a high risk of damage or loss. Additionally, communication limitations arise because meetings are conducted using pre-saved images of lesions during the coordination of pathological findings. To address some of these difficulties, digital pathology has been actively utilized. Over 1,500 papers related to digital pathology have been published [2], and with the advancements in high-resolution scanners and high-performance computers, its practical application has become fully feasible [3]. Whole slide imaging (WSI) in digital pathology refers to the process of scanning glass slides prepared from tissue specimens and storing them in a digital format on a computer. Compared to traditional slide preparation, digital pathology requires additional equipment, skilled personnel, and advanced information technology. Nevertheless, WSI enables diagnosis on a computer screen with a resolution comparable to that of an optical microscope, thereby enhancing accuracy and efficiency. Large-scale digital image data can be securely stored and shared using cloud systems and encryption technologies, facilitating remote diagnosis and collaboration. Additionally, while physical glass slides are challenging to preserve over extended periods, digitized slides offer improved storage and retrieval capabilities [4].
The integration of artificial intelligence (AI) into digital pathology has further advanced the field by enabling automated lesion detection within WSI [5], reducing pathologists’ workloads, and mitigating both interobserver and intraobserver variability [6]. AI encompasses two primary subfields: machine learning and deep learning. Machine learning involves the automatic identification of patterns, situational analysis, and decision-making based on data-driven algorithms [7]. In contrast, deep learning utilizes artificial neural networks or deep neural networks to derive conclusions from data. The primary difference between these technologies lies in the level of human intervention. For example, when an image of a car is input into a neural network for training, the machine extracts features such as wheels, mirrors, and door handles. In machine learning, humans must explicitly guide the process of visual feature extraction. However, in deep learning, the neural network autonomously analyzes data and hierarchically extracts features from low-level to high-level without human guidance. This automated learning process requires larger datasets and more complex models, enabling the learning of intricate patterns. In pathology, object detection technology is widely applied, with deep learning models utilizing convolutional neural networks (CNN) delivering robust performance [8]. The key components of CNNs include convolution layers and pooling layers, which automatically extract essential features from input data. Specifically, convolution layers perform convolution operations across input data using small filters, known as kernels, at regular intervals. This process generates feature maps that capture low-level features like edges and textures and increasingly complex high-level features. By creating diverse feature maps, CNNs effectively learn the intricate structures of images. Pooling layers reduce the size of the feature maps generated by convolution layers by removing unnecessary details while retaining critical features. Pooling operations, such as maximum pooling and average pooling, help eliminate noise and distortions, enhancing model learning efficiency and preventing overfitting. A representative object detection model utilizing these techniques is You Only Look Once (YOLO). The key feature of YOLO is its ability to process an input image in a single network pass to predict both the locations and classes of objects simultaneously [9]. This streamlined structure allows YOLO to perform object detection at high speed. Since its introduction in 2016, YOLO has evolved through multiple versions, from v1 to v8 [10]. The YOLOv8 model used in this study differs from previous YOLO models by employing an anchor-free detection method instead of anchor boxes. Traditional anchor boxes apply pre-defined rectangles of various sizes across image regions to predict object locations. While effective, this approach incurs high computational costs and suffers from reduced accuracy when object sizes differ significantly from the pre-defined anchors. In contrast, the anchor-free method adopts a center-based approach, predicting four distances around the object’s center point to determine object size and class information [11, 12]. Additionally, the YOLOv8 model employs a one-stage object detection method, which performs object localization and classification simultaneously rather than in separate steps, thereby enabling faster processing. Due to these advantages, such as reduced computational complexity and improved accuracy in detecting objects of various sizes and shapes, the YOLOv8 model was considered suitable for detecting apoptotic bodies, which are numerous and vary in size [13]. Our previous studies have also demonstrated the effective use of YOLO models for detecting various renal lesions in digital pathology images. Byun et al. and Bae et al. applied YOLO models (v4 and v8, respectively) to detect renal lesions, achieving high detection performance including up to 98.62% accuracy and notable improvements in diagnostic precision, even for complex, multi-lesion images [14, 15]. Apoptotic bodies are vesicles generated during the process of apoptosis, a form of programmed cell death. Apoptosis, or “programmed cell death,” was first described by Kerr in 1972 [16]. Apoptosis is a form of cell death that occurs naturally during development and aging, maintaining cellular homeostasis in living organisms. There are three representative pathways of apoptosis: the extrinsic pathway, the intrinsic pathway, and the perforin/granzyme pathway, all of which ultimately generate apoptotic bodies, which are then phagocytosed by neighboring cells or macrophages [17]. In pathology, apoptosis is often compared with necrosis, another major type of cell death. Morphologically, apoptosis and necrosis must be clearly differentiated under optical microscopy. Necrosis causes cellular swelling and nuclear changes such as pyknosis, karyorrhexis, and karyolysis. In addition, necrosis is accompanied by inflammation and leakage of cellular components. In contrast, apoptosis involves cellular shrinkage, no release of cellular components, and no inflammatory response.
Apoptotic bodies form through nuclear condensation, DNA fragmentation, and cellular fragmentation, which are rapidly phagocytosed by macrophages or adjacent cells [16]. In H&E-stained tissue slides, apoptotic bodies appear as round or oval structures with condensed cytoplasm or nuclei, creating a hollow space that aids in their identification. Apoptotic bodies typically range from 1–5 μm in size [18, 19], requiring high magnification for microscopic observation. When more than ten apoptotic bodies are found within a single tissue, accurately analyzing these small structures can become time-consuming. Therefore, this study aims to automate the detection of apoptotic bodies using the YOLOv8 model, which is both fast and accurate, to significantly reduce analysis time [20]. By applying the latest digital pathology technologies and the YOLOv8 model, this study seeks to automate the detection of apoptotic bodies in rat liver tissue and analyze its accuracy and efficiency.
MATERIALS AND METHODS
In this study, a 13-week repeated dose toxicity test was conducted, and a total of 478 images of rat livers containing apoptotic bodies were collected from Biotoxtech (Cheongju, Korea), a non-clinical toxicology testing institution in South Korea. Additionally, 46 WSIs of rat livers were obtained from the Animal Resource Bank, Ministry of Food and Drug Safety (Cheongju, Korea). The WSIs were magnified to approximately 40 × using CaseViewer, and 1,080 images containing apoptotic bodies were extracted from the WSIs, resulting in a total of 1,558 images for the study.
Labeling is the process of preparing data so that the model can accurately learn the location and class of objects when training on images. All 1,559 images were labeled using the Roboflow platform.
Data augmentation involves increasing the number and diversity of labeled images using various techniques. In this study, two data augmentation methods were applied: flip (horizontal and vertical symmetry) and rotation. These techniques were chosen to allow the model to recognize objects from different orientations and angles. The flip technique mirrors the images horizontally or vertically to allow the model to recognize objects regardless of their orientation, while the rotation technique allows the model to learn to recognize objects from various angles. After augmentation, the number of images increased to 3,738. This process enables the model to learn various patterns of the target objects, helps prevent overfitting, and improves generalization performance.
The augmented images were divided into a training set, validation set, and test set for the training process. The training set is used to enable the model to learn object patterns and to refine its weights during the training process. The validation set is used during training to evaluate the model’s performance, while the test set is used to assess the final performance of the trained model.
RESULTS
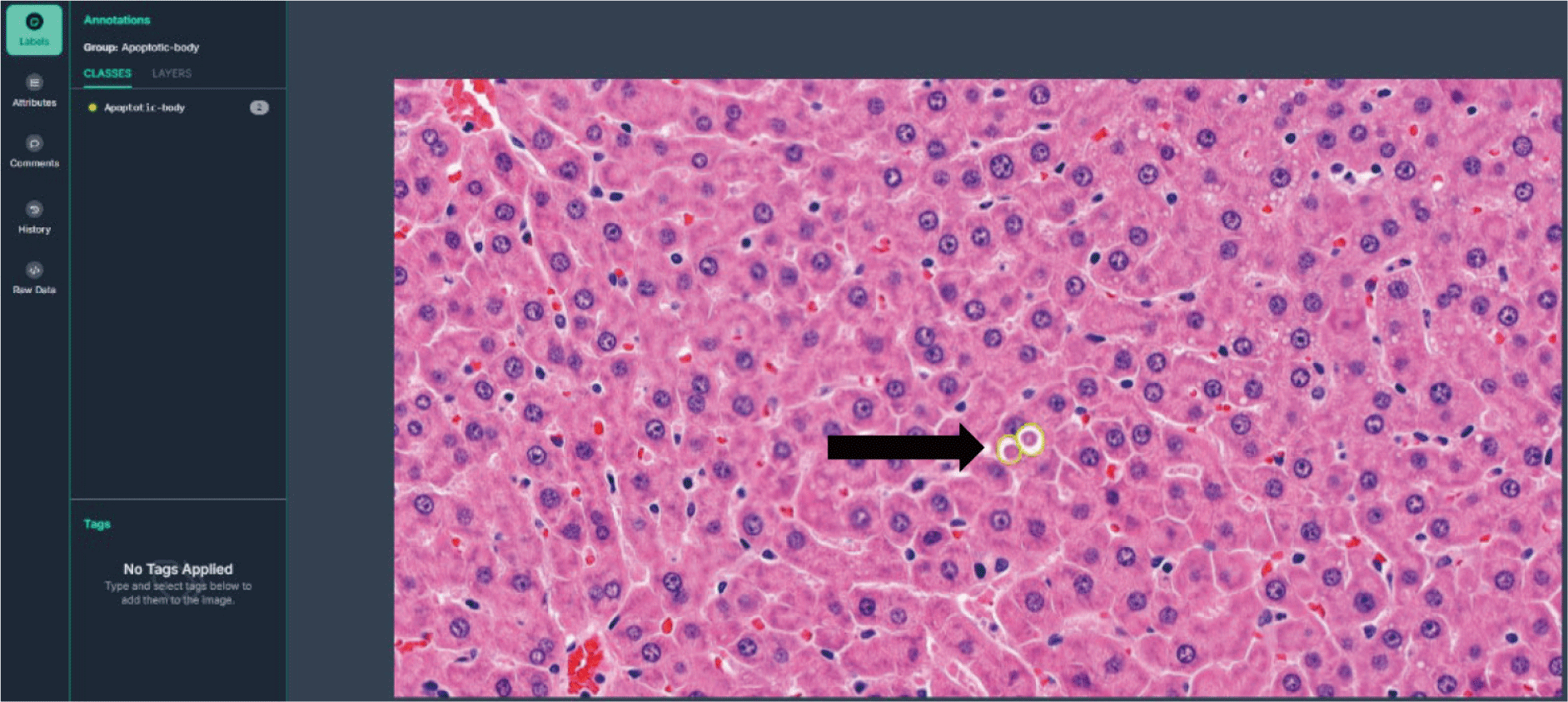
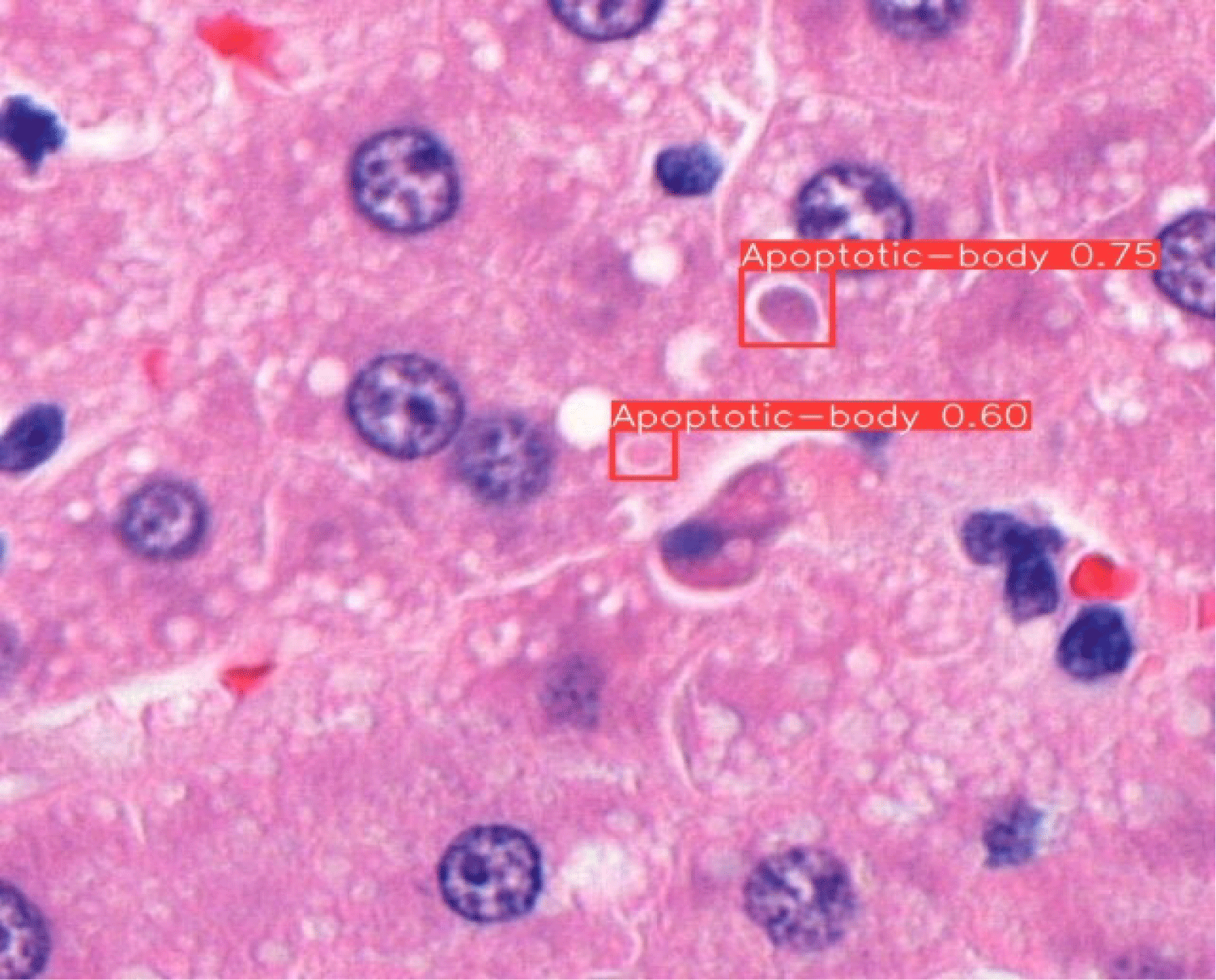
Fig. 1 illustrates the process of annotating apoptotic bodies using the Roboflow platform. The apoptotic bodies (thick arrows) were manually labeled based on morphological characteristics observed in H&E-stained rat liver tissue. Fig. 2 presents the object detection results of apoptotic bodies in H&E-stained rat liver tissue using the YOLOv8 model. The detected apoptotic bodies are shown within red bounding boxes, with confidence scores (e.g., 0.75, 0.60) displayed above each box. The model demonstrates its ability to detect small apoptotic structures with high precision in histopathological imaging.
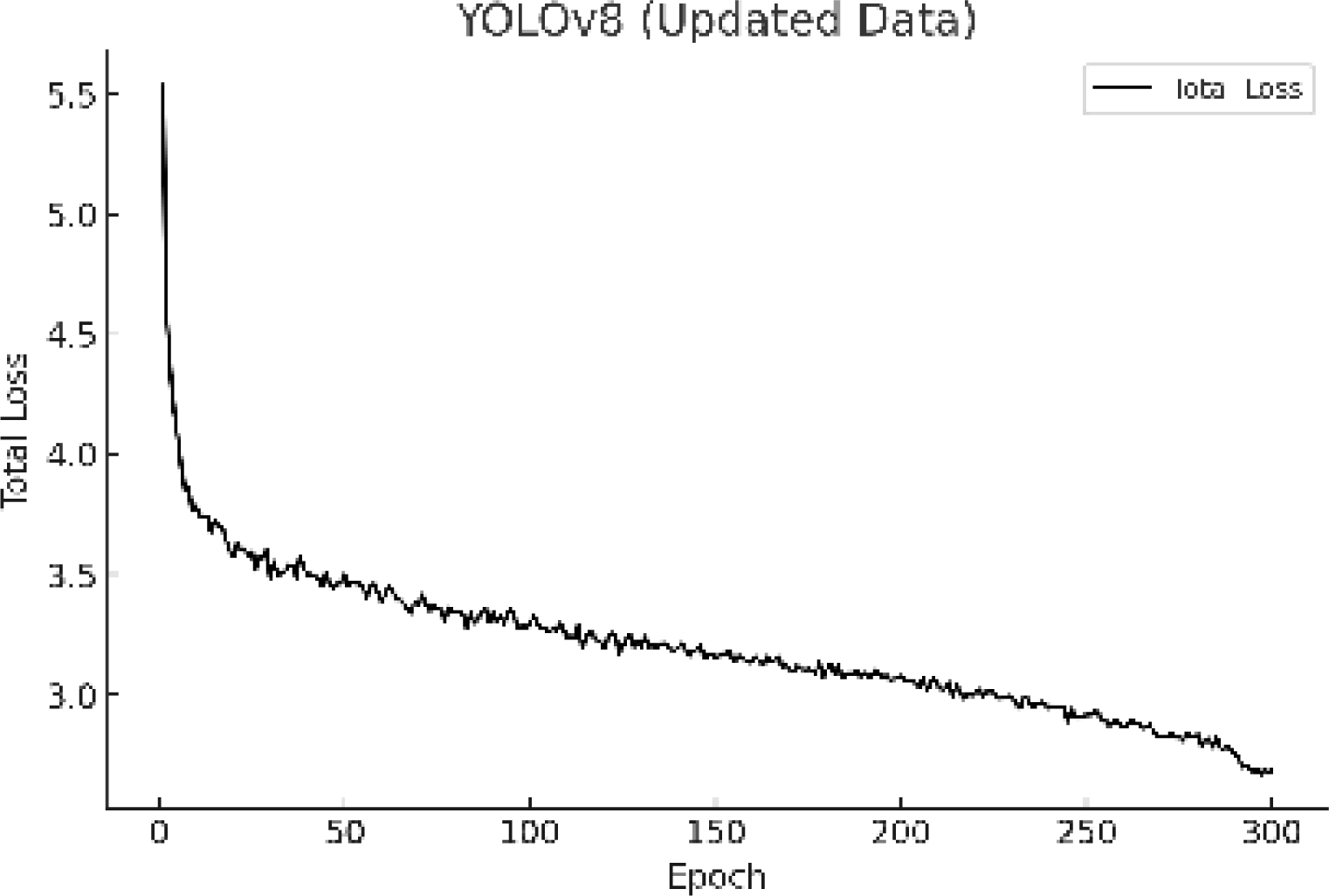
The total loss value was calculated based on the fundamental loss components of the YOLOv8 model, namely localization, classification, and objectness losses. This metric was used to evaluate whether the model was learning effectively prior to assessing its detection accuracy. As shown in Fig. 3, the total loss consistently decreased as training progressed. A steep decline occurred during the initial epochs, followed by a more gradual reduction, suggesting stable convergence of the model. To prevent overfitting, the training process was limited to 300 epochs.
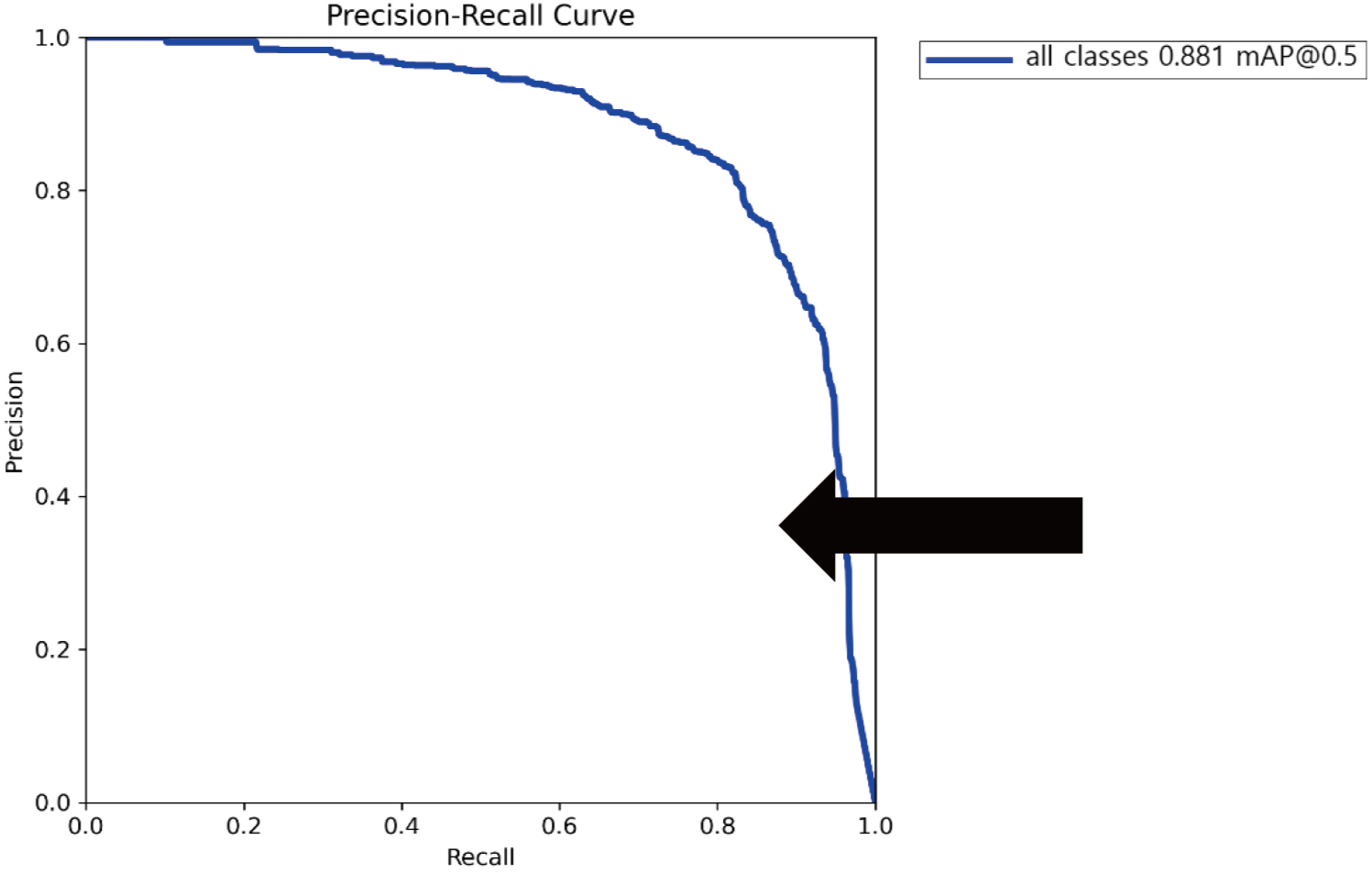
The Precision-Recall curve visualizes the relationship between precision and recall. The Area Under the Curve (AUC; thick arrow) calculated from this graph represents the mean Average Precision (mAP@0.5), a key metric for evaluating the overall performance of object detection. Here, mAP@0.5 refers to the average precision at an Intersection over Union (IoU) threshold of 0.5, which signifies the overlap between the predicted bounding boxes and the ground truth bounding boxes. As shown in Fig. 4, the YOLOv8 model achieved a mAP@0.5 of 0.881 in detecting apoptotic bodies. In addition to the mAP@0.5 value, the model achieved a precision of 1.00, a recall of 0.97, and an F1 score of 0.82, further confirming its robust performance in detecting apoptotic bodies.
DISCUSSION
In this study, the YOLOv8 model demonstrated an accuracy of approximately 88% in detecting apoptotic bodies, confirming its potential as a tool for enhancing digital pathology workflows. The model showed strong performance in terms of precision (1.00), recall (0.97), F1 score (0.82), and mean Average Precision (mAP) of 0.881. These results indicate that YOLOv8 is highly effective for identifying apoptotic bodies in H&E-stained rat liver tissue, supporting its practical application in pathological diagnosis.
The automation of apoptotic body detection through deep learning significantly reduces the workload of pathologists. Traditionally, the identification of apoptotic bodies requires high magnification and is time-consuming. In contrast, the YOLOv8 model enables rapid and accurate detection, streamlining the diagnostic process. By integrating this model into the digital pathology pipeline, pathologists can perform quicker slide reviews, thereby improving both efficiency and diagnostic reliability.
Despite its success, the model still faces challenges in differentiating between apoptotic bodies and other small lesions, such as inclusion bodies, which share similar visual characteristics. The quality of data played a crucial role in achieving the model’s performance. Initially, a dataset of 1,000 images was used, and the model achieved an accuracy of 83.8%. With the addition of 558 more images, the final dataset consisted of 1,558 labeled images, leading to a performance increase to 88.1%. The use of data augmentation further enhanced detection performance by expanding the training data to 3,738 images. These findings underscore the importance of both data quantity and diversity in training deep learning models for medical image analysis.
Small object detection remains a persistent challenge in computer vision, particularly in pathology where critical features like apoptotic bodies are minute and sometimes ambiguous. However, advancements in deep learning—especially object detection algorithms such as YOLOv8—have improved the model’s capability to localize and classify small features. With continued improvements, it is anticipated that the accuracy of such models could exceed 95%, especially when trained with high-resolution data such as WSIs from diverse tissue types.
YOLOv8 has demonstrated excellent performance in apoptotic body detection, particularly in whole-slide image analysis, where its one-stage detection enables fast inference and high computational efficiency. In contrast, two-stage detection models such as Faster R-CNN and Mask R-CNN typically achieve higher localization accuracy, but their multi-step pipelines result in slower inference time. The YOLO series is currently available up to version 11, and future iterations are expected to further improve detection accuracy. These advancements are likely to enhance the practical utility of YOLO-based approaches, providing an improved balance between speed and accuracy in pathological image analysis.
Labeling quality is another critical factor influencing model accuracy. In this study, all annotations were manually created using the Roboflow platform. Although manual labeling ensures high accuracy, it is labor-intensive and can become inconsistent when many small objects are present in a single image. This inconsistency may directly affect the learning process of the model. To address this, future research should explore automated labeling methods, possibly integrating Natural Language Processing (NLP) techniques to generate consistent annotations. Automating the annotation process would significantly improve both efficiency and scalability of training data preparation.
AI-based technologies have been increasingly applied to support lesion detection in toxicopathology. Numerous studies have demonstrated that deep learning–based AI systems can enhance diagnostic efficiency and reduce interobserver variability in toxicological assessments. In this context, the integration of AI approaches such as YOLOv8 is consistent with current technological trends and is expected to facilitate the development of automated and reproducible digital toxicopathology workflows.
Apoptosis plays a critical role in various disease models, including liver toxicity, neurodegenerative disorders, immune-mediated injuries, and chemotherapy-induced tissue damage. Accordingly, automated detection and quantification of apoptotic bodies is expected to provide both research and clinical utility across these disease models.
The practical implications of this study are notable. The developed model has potential to assist pathologists by providing preliminary screenings of digital slides, enabling them to focus more on complex diagnostic tasks. Additionally, by integrating AI-based detection systems into real clinical workflows, the overall speed and accuracy of diagnoses can be enhanced. For broader clinical applicability, future work should aim to validate the model across multiple lesion types and various organ tissues, as well as to develop comprehensive datasets that represent diverse pathological conditions.