Introduction
Human Norovirus (NoV) is a major cause of acute human non-bacterial gastroenteritis worldwide and is characterized by high contagiousness, a short incubation period, and fecal-oral transmission [1-2]. These viruses are members of the family Caliciviridae, genus Norovirus and are divided into six genogroups (GI-GVI) based on the amino acid sequence of the major capsid protein [3-4]. The NoV genome consists of a ~7.5 kb single-stranded, positive-sense RNA containing three open reading frames (ORFs). ORF1 encodes six non-structural proteins (NSP) for viral replication. The ORF2 encodes major capsid protein VP1 while ORF3 encodes the minor structural protein VP2. VP1 is divided into the shell (S) domain and protruding (P) domain. The S domain forms the internal structural core of the particle while the P domain is exposed on the outer surface of the particle. The P domain is further subdivided into the P1 and P2 subdomains. The P2 subdomain is the most exposed surface of the viral particle and is involved in cellular histo-blood group antigen (HBGA) binding and immune reactivity [5].
Since Human NoV (HuNoV) is uncultivable, there are several restrictions in studying this virus. For HuNoV diagnosis, electron microscopy (EM), conventional reverse transcriptional-PCR (RT-PCR), real-time RT-PCR, and enzyme-linked immunosorbent assays (ELISA) have been used [6]. EM is a relatively low sensitive method, requiring special skill and equipment. In general, conventional RT-PCR or real-time RT-PCR is commonly used for NoV detection and diagnosis [7]. Fecal immunological tests such as ELISA and immunochromatographic (ICG) test are technically simple, rapid, and useful for the detection of NoV outbreaks and large-scale tests. However, these assays are less sensitive than PCR. Considering both sensitivity and specificity, RT-PCR has become the gold standard test for diagnosis of NoV [8].
In this study, human NoV VP1 was expressed using baculovirus expression systems, and monoclonal antibodies (MAbs) against NoV VP1 were produced and characterized.
Materials and Methods
A primer pair set containing BamH I and BstB I restriction enzyme sites (underline) was used to amplify the VP1 gene of HuNoV. The sense primer was VP1-F (5’-GATGGATCCTCATGAAGATGGCGTCGAATGA-3’) while the antisense primer was VP1-R (5’-ATCTTCGAAGTTAATGCACGTCTACGCCCCGTT-3’). The coding regions of HuNoV VP1 gene were amplified from viral RNA extracted from the fecal sample, which was confirmed as human Norovirus genogroup II (HuNoV/GII.4) positive by RT-PCR. Amplification of the VP1 gene was performed by RT-PCR at 45°C for 40 min (reverse transcription), 95°C for 15 min (inactivation), 35 cycles of 94°C for 1 min (denaturation), 58°C for 40 sec (annealing), and 72°C for 1 min (extension), followed by 72°C for 5 min (final extension). The PCR products were examined with 1 Kb DNA ladder (Invitrogen) on 1% agarose gel containing ethidium bromide.
The amplified fragments were digested with restriction enzymes BamH I and BstB I (Takara), purified using the ExpinTM Gel SV kit (GeneAll), and then inserted into corresponding regions of pBlueBac4.5/V5-His-TOPO® vector (Invitrogen) containing V5 epitope using T4 DNA ligase. The ligation mixture was transformed into competent cells (HIT DH5-α, RBC) by the heat shock method and grown on a LB agar plate with ampicillin (50 μg/mL). Recombinant vectors were collected from colonies and were checked for target gene insertion by restriction enzyme. Recombinant baculoviruses (rHuNoV/GII) were generated using a Bac-N-blueTM transfection kit (Invitrogen) according to the manufacturer’s protocol. Each recombinant baculovirus was selected and plaque-purified using X-gal. To confirm the presence of the insertion gene in recombinant baculoviruses, PCR was performed using HuNoV VP1 and baculovirus-specific primers. Expressed HuNoV VP1 in recombinant viruses was confirmed by IFA using commercial anti-V5 specific antibody (Bethyl Laboratories) and FITC-conjugated goat anti-rabbit IgG + M (ImmunoResearch Lab). In addition, to identify the expressed NoV VP1 in recombinant viruses, Western blotting was performed by the standard method using anti-V5 specific antibody and alkaline phosphatase-conjugated goat anti-rabbit IgG + M (ImmunoResearch Lab).
HuNoV VP1-specific MAbs were produced using expressed HuNoV VP1 and characterized by using various recombinant viral proteins. For production of HuNoV VP1-specific MAbs, BALB/c mice were immunized with the rHuNoV-VP1-infected Sf9 cell lysates. Cell lysates were emulsified in an equal volume of Freund’s complete adjuvant (Sigma) and then inoculated into each foot pad of a 7-week-old female BALB/c mouse (Samtako Biokorea, Korea). The inoculation was repeated two times every 3 days. After 3 days following final inoculation, popliteal lymph nodes and spleen were removed aseptically from the immunized mouse. The lymphocytes and splenocytes were fused with SP2/0 myeloma cells according to the modified procedures of a previous study [9]. The positive hybridomas were screened by IFA test and cloned at least twice by limiting dilution methods. The isotype of each MAb was determined by a monoclonal antibody isotyping kit (Sino Biological) according to the manufacturer protocol.
Reaction of produced MAbs with other non-human caliciviruses such as porcine Sapovirus, bovine Calicivirus, bovine NoV, and feline Calicivirus was determined by IFA test using the previously expressed recombinant baculoviruses [10]. To determine the reactivity patterns of MAbs with various HuNoV virus-like particles (VLPs), ELISA was used. Briefly, 96-well plates were coated overnight at 4°C with 5 μg/mL of different HuNoV VLPs {GI.1, GI.3, GI.4, GII.3, GII.4 (Asia 2003), GII.4 (Den-Haag 2006b), GII.4 (New-Orleans 2009), GII.4 (Sydney 2012), GII.5, GII.8, GII.10, GII.14, GII.16, and GII.17}, which were kindly provided by the Division of Vaccine Research, Center for Infectious Disease, Korea National Institute of Health. Each well was blocked with 2% skim milk in TBS-T for 2 hr at 37°C. After washing with TBS-T, 1:10 diluted MAbs were added to each well and incubated for 2 hr at 37°C. Following washing six times with TBS-T, 1:5000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG + M (Thermo) was added to each well and incubated for 2 hr at 37°C. After washing six times with TBS-T, 50 μL of TMB (Sigma) was added to the wells and incubated for 15 min at RT to develop the ELISA. The substrate reaction was stopped by adding 50 μL of 1 M H2SO4, and the optical density (OD) was measured at 450 nm.
Results
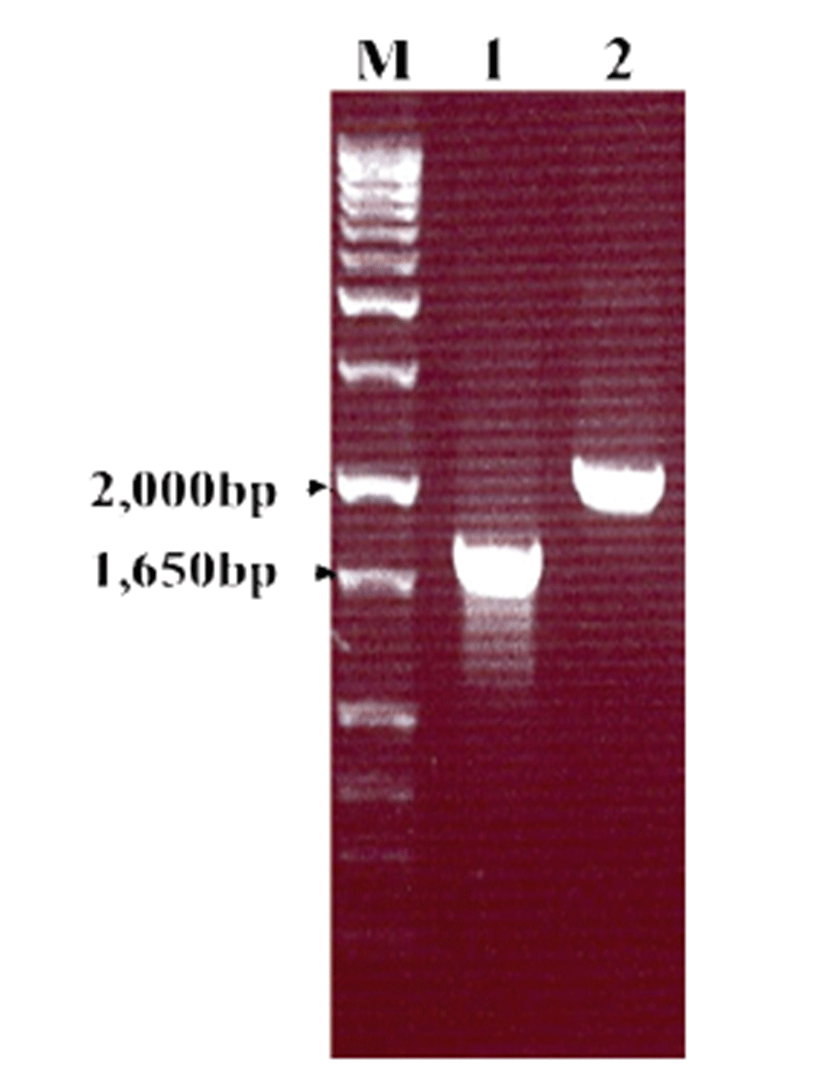
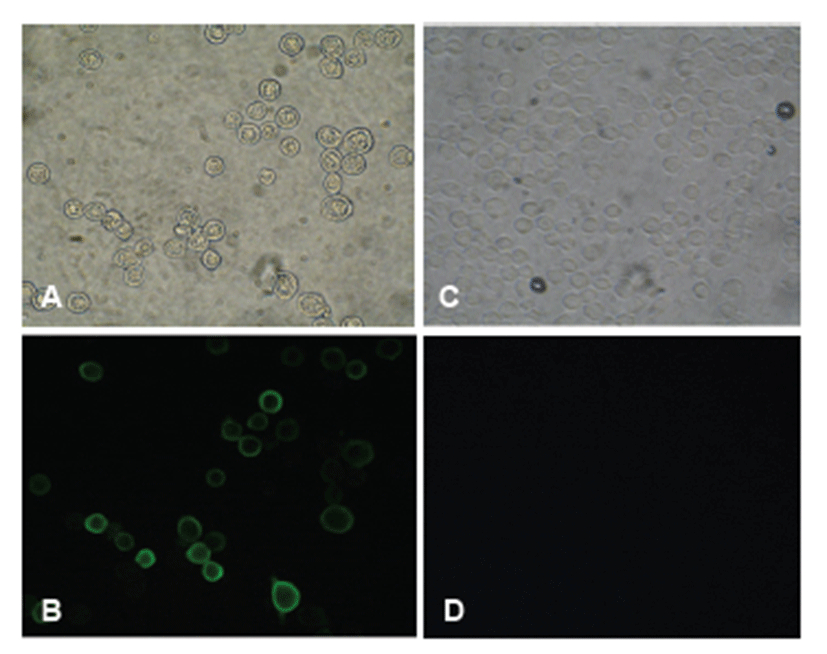
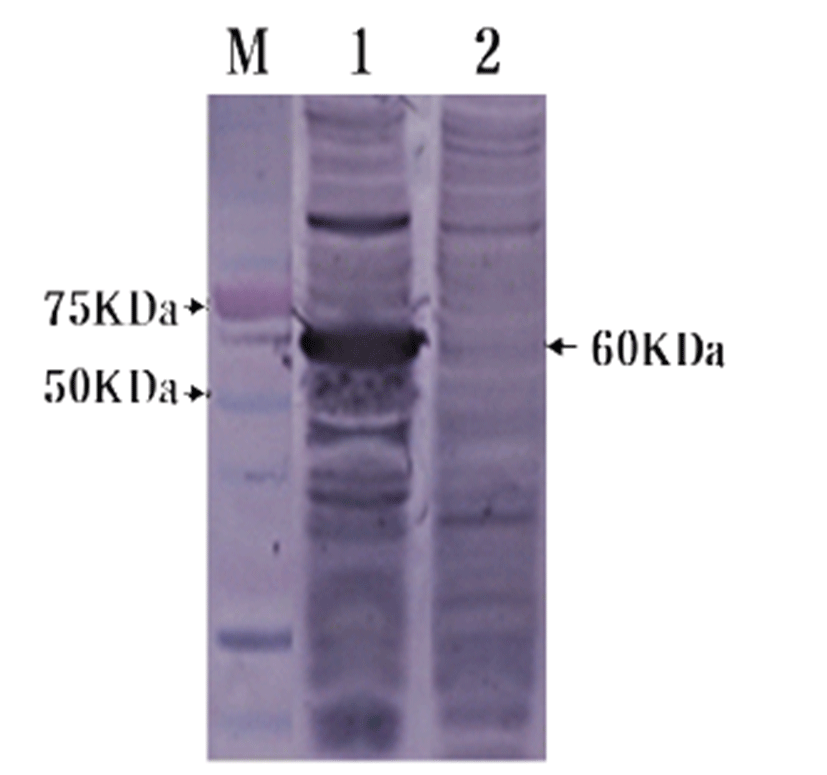
To confirm HuNoV VP1 gene in the recombinant baculovirus (rHuNoV/GII), PCR was performed using HuNoV VP1 and the baculovirus-specific primer set. In this experiment, amplified DNA of expected sizes (1,623 bp and 1,726 bp, respectively) was identified (Fig. 1). Sf9 cells infected with rHuNoV-VP1 showed specific cytopathic activity and were shown to be IFA-positive using anti-V5-specific antibody. There were no reactions in mock-infected Sf9 cells (Fig. 2). As expected, Western blotting showed the presence of a 60 kDa protein band in Sf9 cell lysate infected with rHuNoV/GII (Fig. 3).
The isotype of three MAbs (1F11, 1H8, and 2G10) was identified to be IgG3. The isotype of two MAbs (5D2 and 5G10) was IgM, whereas 4B8 and 5F12 were identified as IgG2b (Table 1). Antigenic specificity of MAbs was confirmed by IFA using GI and GII HuNoV recombinant baculoviruses (rHuNoV/GI and rHuNoV/GII), non-human Calicivirus recombinant baculoviruses (rBoNoro, rPoSapo, and rNB-3), and feline Calicivirus (FCV). All MAbs were identified as GII HuNoV-specific and did not react with non-human Caliciviruses (Table 1). The reactivity patterns of MAbs against different genotypes of HuNoV VLPs were examined by ELISA. All MAbs reacted with only GII.4 genotype. MAb 1F11, 1H8, 2G10, and 5F12 were specific for HuNoV/GII.4 (Asia 2003), HuNoV/GII.4 (Den-Haag 2006b), and HuNoV/GII.4 (New-Orleans 2009) VLPs. MAb 4B8, 5D2, and 5G10 were specific for HuNoV/GII.4 (Den-Haag 2006b) and HuNoV/GII.4 (New-Orleans 2009) VLPs. MAb 1H7 was specific for HuNoV/GII.4 (Den-Haag 2006b) and HuNoV/GII.4 (Sydney 2012) VLPs (Table 2). None of the MAbs reacted with GI genotypes (GI.1, GI.3, and GI4) and other GII genotypes (GII.3, GII.5, GII.8, GII.10, GII.14, GII.16, and GII.17).
rHuNoV/GI: genogroup I human Norovirus recombinant baculovirus, rHuNoV/GII: genogroup II human Norovirus recombinant baculovirus, rBoNoro: bovine Norovirus recombinant baculovirus, rPoSapo: porcine Sapovirus recombinant baculovirus, rNB-3: bovine Calicivirus NB-3 recombinant baculovirus, FCV: native feline Calicivirus.
Discussion
Noroviruses are major viral pathogens causing acute gastroenteritis worldwide [11]. Antigenic diversity and lack of viral cultivation of HuNoV have impaired characterization of HuNoV and development of effective vaccines [12-13]. To overcome this problem, development of vaccines with expressed viral proteins that represent different genotypes has been explored [14]. Baculovirus expression system is widely used for viral protein expression, and numerous recombinant proteins such as NoV VP1 have been produced using this system [15-17]. A unique characteristic of expressed NoV capsid protein VP1 is its ability to self-assemble and form empty virus-like particles (VLPs) [15]. According to previous studies, recombinant NoV VP1 proteins using baculovirus expression systems are known to form self-assembled VLPs composed of 90 copies of major capsid protein dimers. NoV VLPs are structurally and antigenically similar to native NoV particles [18]. For this reason, VLPs are used as antigen substitutes for NoV for studies and development of vaccines. Further, VLPs from expressed NoV capsid proteins are alternative antigens for immunogenicity study [14, 19].
In this study, GII HuNoV VP1 gene was expressed in a baculovirus expression system using recombinant VP1 and GII NoV VP1-specific MAbs. However, this study did not determine whether or not expressed VP1 of HuNoV self-assembled into VLPs. IFA and Western blotting analysis using commercial anti-V5 specific antibody confirmed that the GII HuNoV VP1 gene was successfully expressed in the baculovirus expression system. Immunogenicity of VLPs has been reported in small laboratory animals and non-human primates [19]. Furthermore, several human clinical trials using NoV VLPs have reported immunogenicity of monovalent or bivalent VLPs formulations in adult volunteers [14, 20-21].
There have been several reports on human NoV VP1-specific MAbs. Yoda et al [22, 23] produced MAbs using E. coli-expressed human NoV (GII) recombinant whole capsid protein and reported that some MAbs reacted not only to GII but also to GI recombinant proteins. Li et al [24] produced and characterized MAbs using human NoV GII/strain and found that MAb N2C3 could detect both human and animal-associated NoV. Parra et al [25] reported the characterization of a broadly cross-reactive MAbs raised against VP1 of GII.3 human NoV and found that this MAb reacted with GI, GII, GIV, and GV human NoV. Kou et al [26] characterized broadly cross-reactive MAbs using virus-like particles. For rapid detection of human NoV infection by ELISA, broadly reactive MAbs are needed. However, even though the eight MAbs produced in our studies did not react with non-human Caliciviruses such as bovine NoV, porcine Sapovirus, and feline Calicivirus, we were unable to produce broadly reactive MAbs against HuNoV/GI and HuNoV/GII. In a previous seroepidemiology study, GIII bovine NoV antibodies are detected in human serum most likely due to the cross-reactive epitope between bovine and human NoV [27, 28]. However, MAbs obtained in this study did not bind with rBoNoro.
HuNoV GII is the most prevalent genogroup worldwide, but HuNoV infections due to other genogroups are also important. For this reason, broadly cross-reactive MAbs among different genogroups and genotypes are valuable reagents for the diagnosis and study of HuNoV. Three reactivity patterns of MAbs with various HuNoV VLPs have been recognized. While four MAbs (1F11, 1H8, 2G10, and 5F12) reacted with HuNoV/GII.4 (Asia 2003), HuNoV/GII.4 (Den-Haag 2006b), and HuNoV/GII.4 (New-Orleans 2009), the three other MAbs (4B8, 5D2, and 5G10) recognized HuNoV/GII.4 (Den-Haag 2006b) and HuNoV/GII.4 (New-Orleans 2009). One MAb (1H7) recognized HuNoV/GII.4 (Den-Haag 2006b) and HuNoV/GII.4 (Sydney 2012). From these reactivity patterns, we could assume that at least three epitopes were present in HuNoV VP1. However, whether these epitopes recognized by our MAbs are linear or conformational as well as the exact positions of these epitopes could not be identified, but further studies to determine these problems are in progress.
Ultimately, we obtained recombinant protein of GII HuNoV VP1 gene and at least three different groups of MAbs that specifically bind to different epitopes. VP1 recombinant protein and MAbs developed in this study are expected to be useful for the development of immunological assay such as ELISA and for antigenic mapping of HuNoV VP1 gene.