Introduction
The etiological agents involved in Canine Infectious Respiratory Disease (CIRD) are complex. They are thought to include canine distemper virus (CDV), canine influenza virus (CIV), and canine respiratory coronavirus (CRCoV) [1, 2]. Jeoung et al (2013) reported the multiplex reverse transcription PCR (mRT-PCR) to amplify CRCoV, CIV, and CDV viral target all at the same time, thus detecting multiple respiratory infections within a single sample [3].
A novel coronavirus; canine respiratory CRCoV; was detected in respiratory samples collected from dogs with a high incidence of CIRD in a UK re-homing centre [4], and the role of CRCoV in CIRD is currently under investigation. Coronaviruses are large enveloped viruses containing a single stranded positive-sense RNA genome of approximately 30 kb which encodes a polymerase complex and several structural (spike (S), envelope (E), membrane (M), nucleocapsid (N), and in group two coronaviruses only a haemagglutinin esterase (HE)) and nonstructural proteins. Currently coronaviruses are divided into three groups based on genetic and antigenic traits [5].
Phylogenetic analysis places CRCoV in group 2 of the Coronaviridae family related most closely to bovine coronavirus (BCoV) and human coronavirus OC43 (OC43) [4]. Serological studies have shown CRCoV to be present in the UK, Ireland, Italy, USA and Japan [6-9].
Current detection methods for CRCoV include virus isolation and detection of viral RNA by reverse transcription polymerase chain reaction (RT-PCR) [4, 10]. Cell culture isolation of CRCoV is difficult and time consuming, therefore lacks the sensitivity and speed required for high-throughput screening [4, 8]. Molecular assays such as RT-PCR are faster and offer increased sensitivity and specificity over other detection methods [11, 12], therefore providing the best opportunity for the detection of CRCoV. The RT-PCR method could be available for the detection of CRCoV [4].
In this study, we detected CRCoV infection by a multiplex RT-PCR to amplify CRCoV, CIV, amd CDV viral target from dogs with respiratory disease.
Materials and Methods
Three dogs were obtained from a dog kennel in Iksan City, Korea. Over the previous 5 days, the dogs had shown cough, sneezing, and nasal discharge. All dogs were given health examination. Nasal swabs were collected from the dogs with respiratory symptoms. Samplings were performed from the nares with a dry and unmoistened swab. The tip of the collection swab was inserted into the nares and rolled five times in each nostril. Collected specimens were transported and stored at room temperature. Specimens were processed for RNA extraction within 24 hr of being collected. Each collection swab was put into 2mL of 0.1M PBS buffer and vortexed and discarded, and the PBS was submitted to extract viral RNAs.
The total viral RNA was extracted from each nasal swab sample using the viral RNA kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. First-stand cDNA was synthesized using the first-strand synthesis system with oligo (dT) primer for RT-PCR (Invitrogen, San Diego, CA, U.S.A.).
To detect the etiologic pathogen, we performed a mRT-PCR described by Jeoung et al (2013) to amplify the genes encoding the CIV matrix protein (Accession No: FJ560885), the CDV nucleocapsid protein (Accession No: EF4187830) and the CRCoV spike protein (Accession No: EU983107) [3]. The primers sequences were as follows: CIV MF 5’-TCAGGCCCCCTCAAAGCCG-3’, and CIV MR 5’- CCATCGTCAACATCCACAG C-3’; CRCoV SF 5’- GCAATGCTGGTTCGGAAGAG -3’, and CRCoV SR 5’- GTTGGCAT AGGTGAGCACTG -3’; and CDV NF 5’- CATGGTGGCACTCATCTTGG -3’, and CDV NR 5’-TTCGGACCTCTTGT TGATGG -3’. The multiplex PCR PreMix (Cat. No. K-2111; Bioneer, Daejon, South Korea) was used for mRT-PCR according to the manufacturer’s instructions. 20μL of mRT-PCR reaction mix contained all viral RNA/DNA and all three primer pairs for CIV, CRCoV and CDV in the same tube. The condition for mRTPCR was follows: 95°C for 10 min, followed by 35 cycles of 95°C for 30 sec, 52°C for 30 sec, and 72°C for 60 sec, with a final extension step at 72°C for 5 min and holding at 4°C. Reactions were conducted using My Genie 32 Thermal Block PCR (Bioneer Corporation). Eight microliters of each sample were mixed with 2 μL of loading buffer and electrophoretically separated on 1.2% agarose gels stained with 0.5 μg/mL ethidium bromide. DNA bands were observed under ultraviolet light.
Total viral RNA was extracted from CRCoV-infected cells using the viral RNA kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. cDNA was synthesized using the first-strand synthesis system with oligo (dT) primer (Invitrogen, San Diego, CA, U.S.A.). The template DNA (50 ng) and 20 pmoL of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer Corporation) containing 2.5 U of Taq DNA polymerase, 250 μM each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 40 mM KCl, and 1.5 mM MgCl2. The volume was adjusted with distilled water to 20 μL.
Thereafter, 140 bp fragment of CRCoV nucleocapsid gene was amplified using the primers CRCoV NF3 and CRCoV NR4. The primers CRCoV NF3 (forward 5-CCCTACTATTCTTGGTT-3) and CRCoV NR4 (reverse 5-CGTCTGTTGTGTCTGTACC-3) were designed by Mitchell et al (2009). The reaction mixture was subjected to denaturation at 95°C for 1 min followed by 35 cycles of 95°C for 1 min, 45°C for 40 sec, and 72°C for 1min and a final extension step of 72°C for 10 min. Reactions were conducted using My Genie 32 Thermal Block PCR (Bioneer Corporation).
CRCoV–specific probe was prepared by digoxigenin (DIG)-labeling after amplification by PCR. The PCR products were purified using Wizard PCR preps (Promega, Medison, WI, USA) and then labeled by random priming with DIG-dUTP (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s instructions. Dot blotting was achieved by direct application on a positively charged nylon membrane (Roche Applied Science).
Results
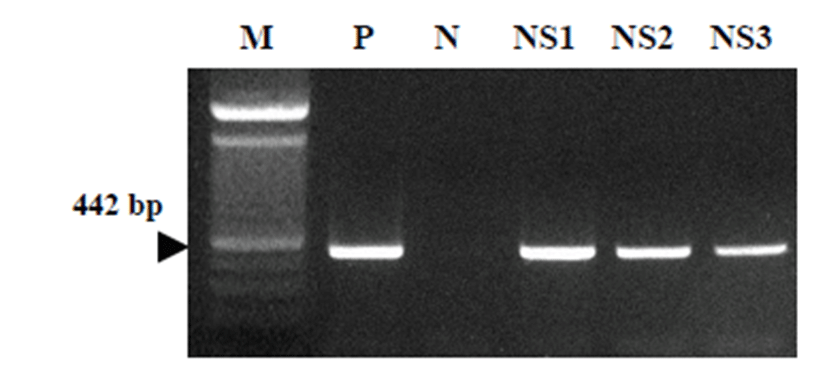
Multiplex RT-PCR with nasal swabbing samples Multiplex RT-PCR was employed to detect to amplify CRCoV, CIV, and CDV viral target. The size of theplified products was 956 bp for CIV, 442 bp for CRCoV, and 624 bp for CDV (Jeong et al, 2013). As the result, the nasal secreting CRCoV was detected by the 442 bp CRCoV positive PCR reaction in the nasal swabbing samples from dogs (Fig. 1).
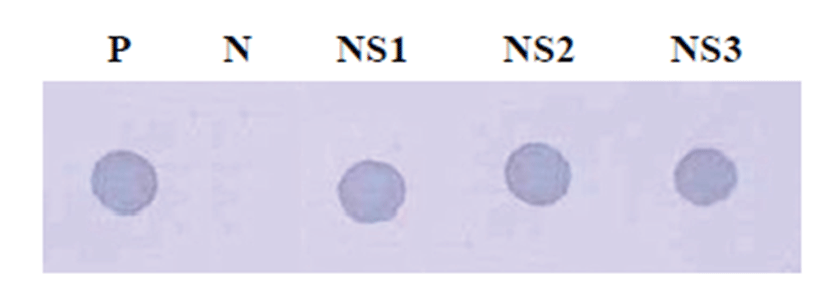
CRCoV-specific probes were prepared by digoxigenin (DIG)-labeling CRCoV nucleocapsid gene amplicon. We performed dot blotting hybridization by direct application on a positively charged nylon membrane with the nasal swab samples. As the results, CRCoV positive reactions were detected in the nasal swabbing samples from dogs (Fig. 2).
Discussion
Viral pathogens are responsible for a significant proportion of the CIRD burden, and new viruses are emerging worldwide. A recent study in South Korea reported CIRD in dogs caused by CIV (H3N2) and CRCoV [13, 14]. CIV H3N2 strain is associated with the development of acute respiratory disease in dogs and is a novel pathogen for CIRD in South Korea and South China [15, 16]. Serological surveys carried out in South Korea, Italy, U.K., Ireland, U.S.A. and Japan, have identified CRCoV as a worldwide pathogen found in the respiratory tract of dogs suffering from mild or severe respiratory disease [3]. CDV is one of the most serious diseases in domestic dogs worldwide [17]. It is highly contagious, affects dogs of all ages, and is associated with high morbidity and mortality [18]. All three viruses are present in South Korea, where 5.1% of dogs are seropositive for CIV and 12.8% are seropositive for CRCoV [13].
This study reports the detection of a canine coronavirus in kenneled dogs with respiratory disease. Coronaviruses have been reported to cause respiratory disease of man, cattle, swine, and poultry, but their presence in the respiratory tract of dogs and a possible association with CIRD has not been determined. The isolations of canine coronavirus from four dogs with symptoms of respiratory disease were reported previously [19]. Therefore it was investigated in this study whether coronaviruses could be detected in dogs from a kennel with CIRD-related symtoms. The disease was endemic in this kennel and could not be controlled by the use of vaccines recommended against “kennel cough,” strongly suggesting that additional agents were contributing to the disease syndrome observed. Samples taken from the respiratory tract of these dogs were examined using RT-PCR primers directed to the conserved polymerase gene of coronaviruses [20].
In this study, we met three dogs with respiratory disease and conducted the health examination. As the results, the dogs with respiratory symptoms revealed CRCoV infection by mRT-PCR assay and dot blot hybridization. mRT-PCR was able to detect successfully CRCoV infection in dogs. We suggested that mRT-PCR would be useful and effective for monitoring CRCoV infection in various kinds of dogs.